Single-cell analysis reveals the spatial-temporal expression of genes associated with esophageal malformations
- PMID: 38355689
- PMCID: PMC10866870
- DOI: 10.1038/s41598-024-53098-w
Single-cell analysis reveals the spatial-temporal expression of genes associated with esophageal malformations
Abstract
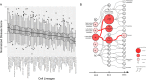
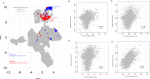
Understanding the molecular mechanisms of congenital diseases is challenging due to their occurrence within specific developmental stages. Esophageal malformations are examples of such conditions, characterized by abnormalities in the development of esophagus during embryogenesis. These developmental malformations encompass a range of anomalies, including esophageal atresia, and tracheoesophageal fistula. Here, we investigated the preferential expression of 29 genes that are implicated in such malformations and their immediate interactome (a total of 67 genes). We conducted our analyses across several single-cell atlases of embryonic development, encompassing approximately 150,000 cells from the mouse foregut, 180,000 cells from human embryos, and 500,000 cells from 24 human organs. Our study, spanning diverse mesodermal and endodermal cell populations and early developmental stages, shows that the genes associated with esophageal malformations show their highest cell-type specific expression in lateral plate mesoderm cells and at the developmental stage of E8.75-E9.0 days. In human embryos, these genes show a significant cell-type specific expression among subpopulations of epithelial cells, fibroblasts and progenitor cells including basal cells. Notably, members of the forkhead-box family of transcription factors, namely FOXF1, FOXC1, and FOXD1, as well as the SRY-box transcription factor, SOX2, demonstrate the most significant preferential expression in both mouse and human embryos. Overall, our findings provide insights into the temporal and cellular contexts contributing to esophageal malformations.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Visualizing expression patterns of Shh and Foxf1 genes in the foregut and lung buds by optical projection tomography.Pediatr Surg Int. 2008 Jan;24(1):3-11. doi: 10.1007/s00383-007-2036-1. Pediatr Surg Int. 2008. PMID: 17962963
-
Altered Tbx1 gene expression is associated with abnormal oesophageal development in the adriamycin mouse model of oesophageal atresia/tracheo-oesophageal fistula.Pediatr Surg Int. 2014 Feb;30(2):143-9. doi: 10.1007/s00383-013-3455-9. Pediatr Surg Int. 2014. PMID: 24356861
-
Genetic factors in esophageal atresia, tracheo-esophageal fistula and the VACTERL association: roles for FOXF1 and the 16q24.1 FOX transcription factor gene cluster, and review of the literature.Eur J Med Genet. 2010 Jan-Feb;53(1):6-13. doi: 10.1016/j.ejmg.2009.10.001. Epub 2009 Oct 12. Eur J Med Genet. 2010. PMID: 19822228 Free PMC article. Review.
-
Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm.Development. 2007 Jul;134(13):2521-31. doi: 10.1242/dev.003855. Epub 2007 May 23. Development. 2007. PMID: 17522155 Free PMC article.
-
[The role of genetic and environmental factors in the etiology of esophageal atresia and tracheo-esophageal fistula].Postepy Hig Med Dosw (Online). 2014 Mar 7;68:238-46. doi: 10.5604/17322693.1093206. Postepy Hig Med Dosw (Online). 2014. PMID: 24662792 Review. Polish.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

