Smoking changes adaptive immunity with persistent effects
- PMID: 38355791
- PMCID: PMC10881394
- DOI: 10.1038/s41586-023-06968-8
Smoking changes adaptive immunity with persistent effects
Abstract
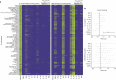
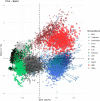
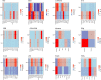
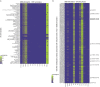
Individuals differ widely in their immune responses, with age, sex and genetic factors having major roles in this inherent variability1-6. However, the variables that drive such differences in cytokine secretion-a crucial component of the host response to immune challenges-remain poorly defined. Here we investigated 136 variables and identified smoking, cytomegalovirus latent infection and body mass index as major contributors to variability in cytokine response, with effects of comparable magnitudes with age, sex and genetics. We find that smoking influences both innate and adaptive immune responses. Notably, its effect on innate responses is quickly lost after smoking cessation and is specifically associated with plasma levels of CEACAM6, whereas its effect on adaptive responses persists long after individuals quit smoking and is associated with epigenetic memory. This is supported by the association of the past smoking effect on cytokine responses with DNA methylation at specific signal trans-activators and regulators of metabolism. Our findings identify three novel variables associated with cytokine secretion variability and reveal roles for smoking in the short- and long-term regulation of immune responses. These results have potential clinical implications for the risk of developing infections, cancers or autoimmune diseases.
© 2024. The Author(s).
Conflict of interest statement
M.L.A. is an employee of Octant Biosciences, CA, USA. D.D. previously received grant support from Rules Based Medicine. The other authors declare no competing interests.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

