Inclusion of deuterated glycopeptides provides increased sequence coverage in hydrogen/deuterium exchange mass spectrometry analysis of SARS-CoV-2 spike glycoprotein
- PMID: 38355883
- PMCID: PMC10871554
- DOI: 10.1002/rcm.9690
Inclusion of deuterated glycopeptides provides increased sequence coverage in hydrogen/deuterium exchange mass spectrometry analysis of SARS-CoV-2 spike glycoprotein
Abstract
Rationale: Hydrogen/deuterium exchange mass spectrometry (HDX-MS) can provide precise analysis of a protein's conformational dynamics across varied states, such as heat-denatured versus native protein structures, localizing regions that are specifically affected by such conditional changes. Maximizing protein sequence coverage provides high confidence that regions of interest were located by HDX-MS, but one challenge for complete sequence coverage is N-glycosylation sites. The deuteration of peptides post-translationally modified by asparagine-bound glycans (glycopeptides) has not always been identified in previous reports of HDX-MS analyses, causing significant sequence coverage gaps in heavily glycosylated proteins and uncertainty in structural dynamics in many regions throughout a glycoprotein.
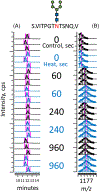
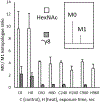
Methods: We detected deuterated glycopeptides with a Tribrid Orbitrap Eclipse mass spectrometer performing data-dependent acquisition. An MS scan was used to identify precursor ions; if high-energy collision-induced dissociation MS/MS of the precursor indicated oxonium ions diagnostic for complex glycans, then electron transfer low-energy collision-induced dissociation MS/MS scans of the precursor identified the modified asparagine residue and the glycan's mass. As in traditional HDX-MS, the identified glycopeptides were then analyzed at the MS level in samples labeled with D2 O.
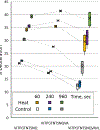
Results: We report HDX-MS analysis of the SARS-CoV-2 spike protein ectodomain in its trimeric prefusion form, which has 22 predicted N-glycosylation sites per monomer, with and without heat treatment. We identified glycopeptides and calculated their average isotopic mass shifts from deuteration. Inclusion of the deuterated glycopeptides increased sequence coverage of spike ectodomain from 76% to 84%, demonstrated that glycopeptides had been deuterated, and improved confidence in results localizing structural rearrangements.
Conclusion: Inclusion of deuterated glycopeptides improves the analysis of the conformational dynamics of glycoproteins such as viral surface antigens and cellular receptors.
Published 2024. This article is a U.S. Government work and is in the public domain in the USA.
Figures
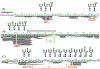


Similar articles
-
Characterization of Site-Specific N- and O-Glycopeptides from Recombinant Spike and ACE2 Glycoproteins Using LC-MS/MS Analysis.Int J Mol Sci. 2024 Dec 20;25(24):13649. doi: 10.3390/ijms252413649. Int J Mol Sci. 2024. PMID: 39769415 Free PMC article.
-
Removal of N-Linked Glycosylations at Acidic pH by PNGase A Facilitates Hydrogen/Deuterium Exchange Mass Spectrometry Analysis of N-Linked Glycoproteins.Anal Chem. 2016 Dec 20;88(24):12479-12488. doi: 10.1021/acs.analchem.6b03951. Epub 2016 Nov 28. Anal Chem. 2016. PMID: 28193043
-
Simultaneous enrichment and separation of neutral and sialyl glycopeptides of SARS-CoV-2 spike protein enabled by dual-functionalized Ti-IMAC material.Anal Bioanal Chem. 2021 Dec;413(29):7295-7303. doi: 10.1007/s00216-021-03433-1. Epub 2021 Jun 21. Anal Bioanal Chem. 2021. PMID: 34155551 Free PMC article.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
-
Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling.Acc Chem Res. 2014 Oct 21;47(10):3018-27. doi: 10.1021/ar500194w. Epub 2014 Aug 29. Acc Chem Res. 2014. PMID: 25171396 Review.
References
-
- Mehra R, Kepp KP. Structure and mutations of SARS-CoV-2 spike protein: A focused overview. ACS Infect Dis. 2022;8(1):29–58. - PubMed
-
- Narang D, James DA, Balmer MT, Wilson DJ. Protein footprinting, conformational dynamics, and core interface-adjacent neutralization “hotspots” in the SARS-CoV-2 spike protein receptor binding domain/human ACE2 interaction. J Am Soc Mass Spectrom. 2021;32(7):1593–1600. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
