Oncoprotein SET-associated transcription factor ZBTB11 triggers lung cancer metastasis
- PMID: 38355937
- PMCID: PMC10867109
- DOI: 10.1038/s41467-024-45585-5
Oncoprotein SET-associated transcription factor ZBTB11 triggers lung cancer metastasis
Abstract
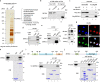
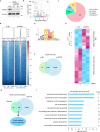
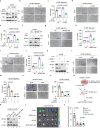
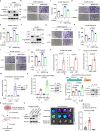
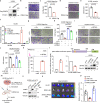
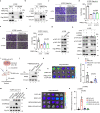
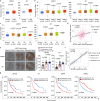
Metastasis is the major cause of lung cancer-related death, but the mechanisms governing lung tumor metastasis remain incompletely elucidated. SE translocation (SET) is overexpressed in lung tumors and correlates with unfavorable prognosis. Here we uncover SET-associated transcription factor, zinc finger and BTB domain-containing protein 11 (ZBTB11), as a prometastatic regulator in lung tumors. SET interacts and collaborates with ZBTB11 to promote lung cancer cell migration and invasion, primarily through SET-ZBTB11 complex-mediated transcriptional activation of matrix metalloproteinase-9 (MMP9). Additionally, by transcriptional repression of proline-rich Gla protein 2 (PRRG2), ZBTB11 links Yes-associated protein 1 (YAP1) activation to drive lung tumor metastasis independently of SET-ZBTB11 complex. Loss of ZBTB11 suppresses distal metastasis in a lung tumor mouse model. Overexpression of ZBTB11 is recapitulated in human metastatic lung tumors and correlates with diminished survival. Our study demonstrates ZBTB11 as a key metastatic regulator and reveals diverse mechanisms by which ZBTB11 modulates lung tumor metastasis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

