Rational Design of a Circularly Permuted Flavin-Based Fluorescent Protein
- PMID: 38356332
- PMCID: PMC11065581
- DOI: 10.1002/cbic.202300814
Rational Design of a Circularly Permuted Flavin-Based Fluorescent Protein
Abstract
Flavin-based fluorescent proteins are oxygen-independent reporters that hold great promise for imaging anaerobic and hypoxic biological systems. In this study, we explored the feasibility of applying circular permutation, a valuable method for the creation of fluorescent sensors, to flavin-based fluorescent proteins. We used rational design and structural data to identify a suitable location for circular permutation in iLOV, a flavin-based reporter derived from A. thaliana. However, relocating the N- and C-termini to this position resulted in a significant reduction in fluorescence. This loss of fluorescence was reversible, however, by fusing dimerizing coiled coils at the new N- and C-termini to compensate for the increase in local chain entropy. Additionally, by inserting protease cleavage sites in circularly permuted iLOV, we developed two protease sensors and demonstrated their application in mammalian cells. In summary, our work establishes the first approach to engineer circularly permuted FbFPs optimized for high fluorescence and further showcases the utility of circularly permuted FbFPs to serve as a scaffold for sensor engineering.
Keywords: biosensing; circular permutation; flavin-based fluorescent proteins; light-oxygen-voltage sensing domain; reporter gene.
© 2024 Wiley-VCH GmbH.
Conflict of interest statement
Conflicts of Interest
There are no conflicts to declare.
Figures
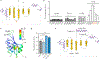
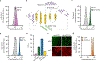
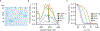
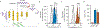
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

