A Case of Okur-Chung Neurodevelopmental Syndrome with a Novel, de novo Variant on the CSNK2A1 Gene in a Turkish Patient
- PMID: 38357263
- PMCID: PMC10862324
- DOI: 10.1159/000530585
A Case of Okur-Chung Neurodevelopmental Syndrome with a Novel, de novo Variant on the CSNK2A1 Gene in a Turkish Patient
Abstract
Introduction: Okur-Chung neurodevelopmental syndrome (OCNDS; #617062) has been associated with heterozygous mutations in the CSNK2A1 gene (*115440) mapped on the chromosome's 20p13 region.
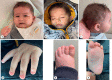
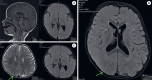
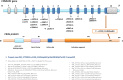
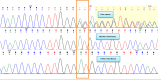
Case presentation: The analysis was performed on a 2-year-old patient who was admitted to our genetic diseases evaluation center by his family with a complaint of hypotonia. We detected a heterozygous NM_177559.3 (CSNK2A1):c.1139_1140dupGG (p.Met381GlyfsTer32) variant in the CSNK2A1 gene from a whole-exome sequence analysis.
Conclusion: The variant that we detected has not been reported in open-access databases to date, so it was evaluated as a novel likely pathogenic variant according to the ACMG-2015 criteria. No variant was detected upon segregation analysis of the patient's parents; therefore, the related variant was evaluated as de novo. In this study, we offer the first report of a pathogenic frameshift variant in the CSNK2A1 gene that has a relationship with OCNDS.
Keywords: De novo variant; Hypotonia; Neurodevelopmental syndrome; Novel; Whole-exome sequencing.
© 2023 S. Karger AG, Basel.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Patient with a heterozygous pathogenic variant in CSNK2A1 gene: A new case to update the Okur-Chung neurodevelopmental syndrome.Am J Med Genet A. 2024 Sep;194(9):e63642. doi: 10.1002/ajmg.a.63642. Epub 2024 May 6. Am J Med Genet A. 2024. PMID: 38711237
-
Clinical Features of Okur-Chung Neurodevelopmental Syndrome: Case Report and Literature Review.Mol Syndromol. 2022 Dec;13(5):381-388. doi: 10.1159/000522353. Epub 2022 Mar 31. Mol Syndromol. 2022. PMID: 36588763 Free PMC article.
-
Dual molecular diagnosis of tricho-rhino-phalangeal syndrome type I and Okur-Chung neurodevelopmental syndrome in one Chinese patient: a case report.BMC Med Genet. 2020 Aug 3;21(1):158. doi: 10.1186/s12881-020-01096-w. BMC Med Genet. 2020. PMID: 32746809 Free PMC article.
-
Patient organization perspective: a research roadmap for Okur-Chung Neurodevelopmental Syndrome.Ther Adv Rare Dis. 2024 Jul 25;5:26330040241249763. doi: 10.1177/26330040241249763. eCollection 2024 Jan-Dec. Ther Adv Rare Dis. 2024. PMID: 39070093 Free PMC article. Review.
-
[A case of Okur-Chung syndrome caused by CSNK2A1 gene variation and review of literature].Zhonghua Er Ke Za Zhi. 2019 May 2;57(5):368-372. doi: 10.3760/cma.j.issn.0578-1310.2019.05.010. Zhonghua Er Ke Za Zhi. 2019. PMID: 31060130 Review. Chinese.
Cited by
-
OCNDS core features are conserved across variants, with loop-region mutations driving greater symptom burden.Front Hum Neurosci. 2025 Jul 3;19:1589897. doi: 10.3389/fnhum.2025.1589897. eCollection 2025. Front Hum Neurosci. 2025. PMID: 40677894 Free PMC article.
-
Adrenal Hypoplasia: A Diagnostic and Clinical Challenge.Cureus. 2025 Jan 27;17(1):e78074. doi: 10.7759/cureus.78074. eCollection 2025 Jan. Cureus. 2025. PMID: 40013223 Free PMC article.
References
-
- de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. . ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362–5. 10.1093/nar/gkl124. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

