The maize striate leaves2 (sr2) gene encodes a conserved DUF3732 domain and is homologous to the rice yss1 gene
- PMID: 38357415
- PMCID: PMC10864124
- DOI: 10.1002/pld3.567
The maize striate leaves2 (sr2) gene encodes a conserved DUF3732 domain and is homologous to the rice yss1 gene
Abstract
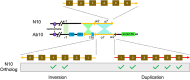
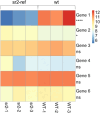
Maize striate leaves2 (sr2) is a mutant that causes white stripes on leaves that has been used in mapping studies for decades though the underlying gene has not been identified. The sr2 locus has been previously mapped to small regions of normal chromosome 10 (N10) and a rearranged variant called abnormal chromosome 10 (Ab10). A comparison of assembled genomes carrying N10 and Ab10 revealed only five candidate sr2 genes. Analysis of a stock carrying the sr2 reference allele (sr2-ref) showed that one of the five genes has a transposon insertion that disrupts its protein sequence and has a severe reduction in mRNA. An independent Mutator transposon insertion in the gene (sr2-Mu) failed to complement the sr2-ref mutation, and plants homozygous for sr2-Mu showed white striped leaf margins. The sr2 gene encodes a DUF3732 protein with strong homology to a rice gene with a similar mutant phenotype called young seedling stripe1 (yss1). These and other published data suggest that sr2 may have a function in plastid gene expression.
Keywords: Zea mays; chloroplast development; comparative genomics; transposon induced mutation.
© 2024 The Authors. Plant Direct published by American Society of Plant Biologists and the Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
We have no conflicts of interest to declare.
Figures


References
-
- Bioinformatics B (2014). FastQC. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

