Deep learning-based predictive model for pathological complete response to neoadjuvant chemotherapy in breast cancer from biopsy pathological images: a multicenter study
- PMID: 38357498
- PMCID: PMC10864440
- DOI: 10.3389/fphys.2024.1279982
Deep learning-based predictive model for pathological complete response to neoadjuvant chemotherapy in breast cancer from biopsy pathological images: a multicenter study
Abstract
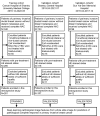
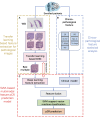
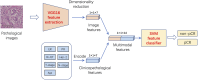
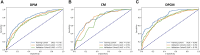
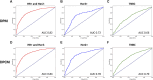
Introduction: Early predictive pathological complete response (pCR) is beneficial for optimizing neoadjuvant chemotherapy (NAC) strategies for breast cancer. The hematoxylin and eosin (HE)-stained slices of biopsy tissues contain a large amount of information on tumor epithelial cells and stromal. The fusion of pathological image features and clinicopathological features is expected to build a model to predict pCR of NAC in breast cancer. Methods: We retrospectively collected a total of 440 breast cancer patients from three hospitals who underwent NAC. HE-stained slices of biopsy tissues were scanned to form whole-slide images (WSIs), and pathological images of representative regions of interest (ROI) of each WSI were selected at different magnifications. Based on several different deep learning models, we propose a novel feature extraction method on pathological images with different magnifications. Further, fused with clinicopathological features, a multimodal breast cancer NAC pCR prediction model based on a support vector machine (SVM) classifier was developed and validated with two additional validation cohorts (VCs). Results: Through experimental validation of several different deep learning models, we found that the breast cancer pCR prediction model based on the SVM classifier, which uses the VGG16 model for feature extraction of pathological images at ×20 magnification, has the best prediction efficacy. The area under the curve (AUC) of deep learning pathological model (DPM) were 0.79, 0.73, and 0.71 for TC, VC1, and VC2, respectively, all of which exceeded 0.70. The AUCs of clinical model (CM), a clinical prediction model established by using clinicopathological features, were 0.79 for TC, 0.73 for VC1, and 0.71 for VC2, respectively. The multimodal deep learning clinicopathological model (DPCM) established by fusing pathological images and clinicopathological features improved the AUC of TC from 0.79 to 0.84. The AUC of VC2 improved from 0.71 to 0.78. Conclusion: Our study reveals that pathological images of HE-stained slices of pre-NAC biopsy tissues can be used to build a pCR prediction model. Combining pathological images and clinicopathological features can further enhance the predictive efficacy of the model.
Keywords: breast cancer; deep learning; neoadjuvant chemotherapy; pathological complete response; pathological images.
Copyright © 2024 Zeng, Qiu, Zhuang, Wei, Wu, Zhang, Chen, Wu and Zhuang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Deep learning with biopsy whole slide images for pretreatment prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer:A multicenter study.Breast. 2022 Dec;66:183-190. doi: 10.1016/j.breast.2022.10.004. Epub 2022 Oct 19. Breast. 2022. PMID: 36308926 Free PMC article.
-
Digital image analysis and machine learning-assisted prediction of neoadjuvant chemotherapy response in triple-negative breast cancer.Breast Cancer Res. 2024 Jan 18;26(1):12. doi: 10.1186/s13058-023-01752-y. Breast Cancer Res. 2024. PMID: 38238771 Free PMC article.
-
[Prediction of Response to Neoadjuvant Chemotherapy for Breast Cancer Based on Histomorphology Analysis of Needle Biopsy Images].Sichuan Da Xue Xue Bao Yi Xue Ban. 2021 Mar;52(2):279-285. doi: 10.12182/20210360505. Sichuan Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 33829703 Free PMC article. Chinese.
-
Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method.Thorac Cancer. 2020 Mar;11(3):651-658. doi: 10.1111/1759-7714.13309. Epub 2020 Jan 16. Thorac Cancer. 2020. PMID: 31944571 Free PMC article.
-
Digital image analysis and machine learning-assisted prediction of neoadjuvant chemotherapy response in triple-negative breast cancer.Res Sq [Preprint]. 2023 Aug 18:rs.3.rs-3243195. doi: 10.21203/rs.3.rs-3243195/v1. Res Sq. 2023. Update in: Breast Cancer Res. 2024 Jan 18;26(1):12. doi: 10.1186/s13058-023-01752-y. PMID: 37645881 Free PMC article. Updated. Preprint.
Cited by
-
Artificial Intelligence-Based Pathology to Assist Prediction of Neoadjuvant Therapy Responses for Breast Cancer.Cancer Med. 2025 Aug;14(15):e71132. doi: 10.1002/cam4.71132. Cancer Med. 2025. PMID: 40762329 Free PMC article. Review.
-
A preoperative predictive model based on multi-modal features to predict pathological complete response after neoadjuvant chemoimmunotherapy in esophageal cancer patients.Front Immunol. 2025 Jan 27;16:1530279. doi: 10.3389/fimmu.2025.1530279. eCollection 2025. Front Immunol. 2025. PMID: 39958355 Free PMC article.
-
Multimodal deep learning for predicting neoadjuvant treatment outcomes in breast cancer: a systematic review.Biol Direct. 2025 Jun 23;20(1):72. doi: 10.1186/s13062-025-00661-8. Biol Direct. 2025. PMID: 40551237 Free PMC article.
-
DLCNBC-SA: a model for assessing axillary lymph node metastasis status in early breast cancer patients.Quant Imaging Med Surg. 2024 Aug 1;14(8):5831-5844. doi: 10.21037/qims-24-257. Epub 2024 Jul 26. Quant Imaging Med Surg. 2024. PMID: 39144041 Free PMC article.
-
Applications of artificial intelligence in digital pathology for gastric cancer.Front Oncol. 2024 Oct 28;14:1437252. doi: 10.3389/fonc.2024.1437252. eCollection 2024. Front Oncol. 2024. PMID: 39529836 Free PMC article. Review.
References
-
- Ali H. R., Dariush A., Thomas J., Provenzano E., Dunn J., Hiller L., et al. (2017). Lymphocyte density determined by computational pathology validated as a predictor of response to neoadjuvant chemotherapy in breast cancer: secondary analysis of the ARTemis trial. Ann. Oncol. 28 (8), 1832–1835. 10.1093/annonc/mdx266 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

