A metabolic signature for NADSYN1-dependent congenital NAD deficiency disorder
- PMID: 38357931
- PMCID: PMC10866660
- DOI: 10.1172/JCI174824
A metabolic signature for NADSYN1-dependent congenital NAD deficiency disorder
Abstract
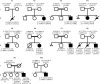
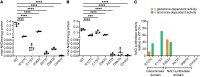
Nicotinamide adenine dinucleotide (NAD) is essential for embryonic development. To date, biallelic loss-of-function variants in 3 genes encoding nonredundant enzymes of the NAD de novo synthesis pathway - KYNU, HAAO, and NADSYN1 - have been identified in humans with congenital malformations defined as congenital NAD deficiency disorder (CNDD). Here, we identified 13 further individuals with biallelic NADSYN1 variants predicted to be damaging, and phenotypes ranging from multiple severe malformations to the complete absence of malformation. Enzymatic assessment of variant deleteriousness in vitro revealed protein domain-specific perturbation, complemented by protein structure modeling in silico. We reproduced NADSYN1-dependent CNDD in mice and assessed various maternal NAD precursor supplementation strategies to prevent adverse pregnancy outcomes. While for Nadsyn1+/- mothers, any B3 vitamer was suitable to raise NAD, preventing embryo loss and malformation, Nadsyn1-/- mothers required supplementation with amidated NAD precursors (nicotinamide or nicotinamide mononucleotide) bypassing their metabolic block. The circulatory NAD metabolome in mice and humans before and after NAD precursor supplementation revealed a consistent metabolic signature with utility for patient identification. Our data collectively improve clinical diagnostics of NADSYN1-dependent CNDD, provide guidance for the therapeutic prevention of CNDD, and suggest an ongoing need to maintain NAD levels via amidated NAD precursor supplementation after birth.
Keywords: Embryonic development; Genetic diseases; Mouse models; Reproductive biology; Therapeutics.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

