The RIN4-like/NOI proteins NOI10 and NOI11 modulate the response to biotic stresses mediated by RIN4 in Arabidopsis
- PMID: 38358510
- PMCID: PMC10869442
- DOI: 10.1007/s00299-024-03151-9
The RIN4-like/NOI proteins NOI10 and NOI11 modulate the response to biotic stresses mediated by RIN4 in Arabidopsis
Abstract
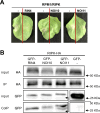
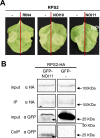
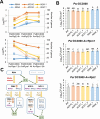
NOI10 and NOI11 are two RIN4-like/NOI proteins that participate in the immune response of the Arabidopsis plant and affect the RIN4-regulated mechanisms involving the R-proteins RPM1 and RPS2. The immune response in plants depends on the regulation of signaling pathways triggered by pathogens and herbivores. RIN4, a protein of the RIN4-like/NOI family, is considered to be a central immune signal in the interactions of plants and pathogens. In Arabidopsis thaliana, four of the 15 members of the RIN4-like/NOI family (NOI3, NOI5, NOI10, and NOI11) were induced in response to the plant herbivore Tetranychus urticae. While overexpressing NOI10 and NOI11 plants did not affect mite performance, opposite callose accumulation patterns were observed when compared to RIN4 overexpressing plants. In vitro and in vivo analyses demonstrated the interaction of NOI10 and NOI11 with the RIN4 interactors RPM1, RPS2, and RIPK, suggesting a role in the context of the RIN4-regulated immune response. Transient expression experiments in Nicotiana benthamiana evidenced that NOI10 and NOI11 differed from RIN4 in their functionality. Furthermore, overexpressing NOI10 and NOI11 plants had significant differences in susceptibility with WT and overexpressing RIN4 plants when challenged with Pseudomonas syringae bacteria expressing the AvrRpt2 or the AvrRpm1 effectors. These results demonstrate the participation of NOI10 and NOI11 in the RIN4-mediated pathway. Whereas RIN4 is considered a guardee protein, NOI10 and NOI11 could act as decoys to modulate the concerted activity of effectors and R-proteins.
Keywords: Arabidopsis; Biotic stress; Plant defense; Pseudomonas syringae; RIN4-like/NOI proteins; Tetranychus urticae.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests to disclose.
Figures
Similar articles
-
Comparative and evolutionary analysis of Arabidopsis RIN4-like/NOI proteins induced by herbivory.PLoS One. 2022 Sep 27;17(9):e0270791. doi: 10.1371/journal.pone.0270791. eCollection 2022. PLoS One. 2022. PMID: 36166429 Free PMC article.
-
RIN4 homologs from important crop species differentially regulate the Arabidopsis NB-LRR immune receptor, RPS2.Plant Cell Rep. 2021 Dec;40(12):2341-2356. doi: 10.1007/s00299-021-02771-9. Epub 2021 Sep 5. Plant Cell Rep. 2021. PMID: 34486076
-
Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1.Plant Cell. 2004 Oct;16(10):2822-35. doi: 10.1105/tpc.104.024117. Epub 2004 Sep 10. Plant Cell. 2004. PMID: 15361584 Free PMC article.
-
Role of RIN4 in Regulating PAMP-Triggered Immunity and Effector-Triggered Immunity: Current Status and Future Perspectives.Mol Cells. 2019 Jul 31;42(7):503-511. doi: 10.14348/molcells.2019.2433. Mol Cells. 2019. PMID: 31362467 Free PMC article. Review.
-
Functions of RPM1-interacting protein 4 in plant immunity.Planta. 2021 Jan 3;253(1):11. doi: 10.1007/s00425-020-03527-7. Planta. 2021. PMID: 33389186 Review.
Cited by
-
Cullin-Conciliated Regulation of Plant Immune Responses: Implications for Sustainable Crop Protection.Plants (Basel). 2024 Oct 26;13(21):2997. doi: 10.3390/plants13212997. Plants (Basel). 2024. PMID: 39519916 Free PMC article. Review.
-
Overexpression of FLZ12 Suppresses Root Hair Development and Enhances Iron-Deficiency Tolerance in Arabidopsis.Genes (Basel). 2025 Apr 6;16(4):438. doi: 10.3390/genes16040438. Genes (Basel). 2025. PMID: 40282398 Free PMC article.
References
-
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. - DOI - PMC - PubMed
-
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

