Senescence-related genes analysis in breast cancer reveals the immune microenvironment and implications for immunotherapy
- PMID: 38358910
- PMCID: PMC10929821
- DOI: 10.18632/aging.205544
Senescence-related genes analysis in breast cancer reveals the immune microenvironment and implications for immunotherapy
Abstract
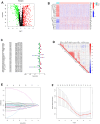
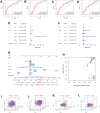
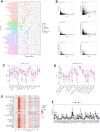
Despite the advent of precision therapy for breast cancer (BRCA) treatment, some individuals are still unable to benefit from it and have poor survival prospects as a result of the disease's high heterogeneity. Cell senescence plays a crucial role in the tumorigenesis, progression, and immune regulation of cancer and has a major impact on the tumor microenvironment. To find new treatment strategies, we aimed to investigate the potential significance of cell senescence in BRCA prognosis and immunotherapy. We created a 9-gene senescence-related signature. We evaluated the predictive power and the role of signatures in the immune microenvironment and infiltration. In vitro tests were used to validate the expression and function of the distinctive critical gene ACTC1. Our risk signature allows BRCA patients to receive a Predictive Risk Signature (PRS), which may be used to further categorize a patient's response to immunotherapy. Compared to conventional clinicopathological characteristics, PRS showed strong predictive efficacy and precise survival prediction. Moreover, PRS subgroups were examined for altered pathways, mutational patterns, and possibly useful medicines. Our research offers suggestions for incorporating senescence-based molecular classification into risk assessment and ICI therapy decision-making.
Keywords: breast cancer; immune microenvironment; immunotherapy; risk signature; senescence.
Conflict of interest statement
Figures


Similar articles
-
Multi-omics analysis revealing a senescence-relevant lncRNAs signature for the assessment of response to immunotherapy for breast cancer.Medicine (Baltimore). 2023 Jul 14;102(28):e34287. doi: 10.1097/MD.0000000000034287. Medicine (Baltimore). 2023. PMID: 37443486 Free PMC article.
-
Oncogenic signaling pathway-related long non-coding RNAs for predicting prognosis and immunotherapy response in breast cancer.Front Immunol. 2022 Aug 4;13:891175. doi: 10.3389/fimmu.2022.891175. eCollection 2022. Front Immunol. 2022. PMID: 35990668 Free PMC article.
-
Comprehensive analysis of cuproptosis-related genes and tumor microenvironment infiltration characterization in breast cancer.Front Immunol. 2022 Oct 20;13:978909. doi: 10.3389/fimmu.2022.978909. eCollection 2022. Front Immunol. 2022. PMID: 36341328 Free PMC article.
-
Spatial transcriptomics: a new frontier in accurate localization of breast cancer diagnosis and treatment.Front Immunol. 2024 Oct 8;15:1483595. doi: 10.3389/fimmu.2024.1483595. eCollection 2024. Front Immunol. 2024. PMID: 39439806 Free PMC article. Review.
-
The current advances of lncRNAs in breast cancer immunobiology research.Front Immunol. 2023 Jun 5;14:1194300. doi: 10.3389/fimmu.2023.1194300. eCollection 2023. Front Immunol. 2023. PMID: 37342324 Free PMC article. Review.
Cited by
-
Senescence-related genes as prognostic indicators in breast cancer survival.Geroscience. 2025 Jun;47(3):2995-3006. doi: 10.1007/s11357-024-01384-w. Epub 2024 Oct 21. Geroscience. 2025. PMID: 39432147 Free PMC article.
-
Cancer-associated fibroblasts gene signature: a novel approach to survival prediction and immunotherapy guidance in colon cancer.Front Immunol. 2025 Apr 8;16:1532306. doi: 10.3389/fimmu.2025.1532306. eCollection 2025. Front Immunol. 2025. PMID: 40264753 Free PMC article.
References
-
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001; 344:783–92. 10.1056/NEJM200103153441101 - DOI - PubMed
-
- Ahn HK, Sim SH, Suh KJ, Kim MH, Jeong JH, Kim JY, Lee DW, Ahn JH, Chae H, Lee KH, Kim JH, Lee KS, Sohn JH, et al.. Response Rate and Safety of a Neoadjuvant Pertuzumab, Atezolizumab, Docetaxel, and Trastuzumab Regimen for Patients With ERBB2-Positive Stage II/III Breast Cancer: The Neo-PATH Phase 2 Nonrandomized Clinical Trial. JAMA Oncol. 2022; 8:1271–7. 10.1001/jamaoncol.2022.2310 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

