Identification of immunogenic cell death-related genes involved in Alzheimer's disease
- PMID: 38360834
- PMCID: PMC10869701
- DOI: 10.1038/s41598-024-54357-6
Identification of immunogenic cell death-related genes involved in Alzheimer's disease
Abstract
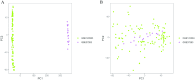
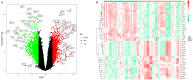
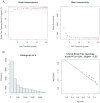
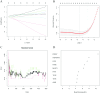
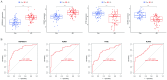
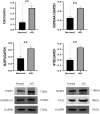
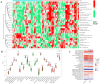
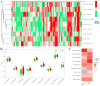
Alzheimer's disease (AD) is the leading cause of dementia worldwide, with recent studies highlighting the potential role of immunogenic cell death (ICD) in the pathogenesis of this neurodegenerative disorder. A total of 52 healthy controls and 64 patients with AD were included. Compared to the controls, the patients with AD exhibited 2392 differentially expressed genes (DEGs), of which 1015 and 1377 were upregulated and downregulated genes, respectively. Among them, nine common genes were identified by intersecting the AD-related module genes with the DEGs and ICD-associated genes. Gene ontology (GO)analysis further revealed "positive regulation of cytokine production" as the most significant term. Moreover, the enriched molecular functions were primarily related to the inflammatory body complex, while the overlapping genes were significantly enriched in lipopolysaccharide binding. Kyoto encyclopedia of genes and genomes (KEGG) analysis also indicated that these overlapping genes were mainly enriched in immunity, inflammation, and lipid metabolism pathways. Furthermore, the following four hub genes were detected using machine learning algorithms: P2RX7, HSP90AA1, NT5E, and NLRP3. These genes demonstrated significant differences in expression between the AD and healthy control groups (P < 0.05). Additionally, the area under the curve values of these four genes were all > 0.7, indicating their potential diagnostic value for AD. We further validated the protein levels of these four genes in the hippocampus of 3xTg-AD and C57BL/6J mice, showing P2RX7 and HSP90AA1 expression levels consistent with the previously analyzed trends. Finally, the single-sample gene set enrichment analysis (ssGSEA) algorithm provided additional evidence by demonstrating the crucial role of immune cell infiltration and its link with the hub genes in AD progression. Our study results suggest that ICD-mediated elevation of HSP90AA1 and P2RX7 levels and the resulting induction of tau hyperphosphorylation and neuroinflammation are vital in the AD pathogenic mechanism.
Keywords: Alzheimer’s disease; GEO; Immunogenic cell death; WGCNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- 2021 Alzheimer's disease facts and figures. Alzheimer's Dement.17, 327–406. 10.1002/alz.12328 (2021). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

