Diversity and evolution of the vertebrate chemoreceptor gene repertoire
- PMID: 38360851
- PMCID: PMC10869828
- DOI: 10.1038/s41467-024-45500-y
Diversity and evolution of the vertebrate chemoreceptor gene repertoire
Abstract
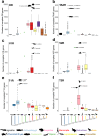
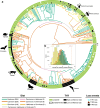
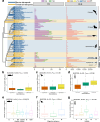
Chemoreception - the ability to smell and taste - is an essential sensory modality of most animals. The number and type of chemical stimuli that animals can perceive depends primarily on the diversity of chemoreceptors they possess and express. In vertebrates, six families of G protein-coupled receptors form the core of their chemosensory system, the olfactory/pheromone receptor gene families OR, TAAR, V1R and V2R, and the taste receptors T1R and T2R. Here, we study the vertebrate chemoreceptor gene repertoire and its evolutionary history. Through the examination of 1,527 vertebrate genomes, we uncover substantial differences in the number and composition of chemoreceptors across vertebrates. We show that the chemoreceptor gene families are co-evolving, highly dynamic, and characterized by lineage-specific expansions (for example, OR in tetrapods; TAAR, T1R in teleosts; V1R in mammals; V2R, T2R in amphibians) and losses. Overall, amphibians, followed by mammals, are the vertebrate clades with the largest chemoreceptor repertoires. While marine tetrapods feature a convergent reduction of chemoreceptor numbers, the number of OR genes correlates with habitat in mammals and birds and with migratory behavior in birds, and the taste receptor repertoire correlates with diet in mammals and with aquatic environment in fish.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Taste and odorant receptors of the coelacanth--a gene repertoire in transition.J Exp Zool B Mol Dev Evol. 2014 Sep;322(6):403-14. doi: 10.1002/jez.b.22531. Epub 2013 Sep 18. J Exp Zool B Mol Dev Evol. 2014. PMID: 24106203
-
Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium.Mol Biol Evol. 2007 Sep;24(9):2099-107. doi: 10.1093/molbev/msm140. Epub 2007 Jul 18. Mol Biol Evol. 2007. PMID: 17634392
-
Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles.Genome Biol Evol. 2013;5(2):389-401. doi: 10.1093/gbe/evt013. Genome Biol Evol. 2013. PMID: 23348039 Free PMC article.
-
Extraordinary diversity of chemosensory receptor gene repertoires among vertebrates.Results Probl Cell Differ. 2009;47:1-23. doi: 10.1007/400_2008_4. Results Probl Cell Differ. 2009. PMID: 19145414 Review.
-
Vomeronasal Receptors in Vertebrates and the Evolution of Pheromone Detection.Annu Rev Anim Biosci. 2017 Feb 8;5:353-370. doi: 10.1146/annurev-animal-022516-022801. Epub 2016 Nov 28. Annu Rev Anim Biosci. 2017. PMID: 27912243 Review.
Cited by
-
Type 2 vomeronasal receptor expression in the olfactory organ of African lungfish, Protopterus annectens.Cell Tissue Res. 2024 Nov;398(2):79-91. doi: 10.1007/s00441-024-03918-2. Epub 2024 Sep 30. Cell Tissue Res. 2024. PMID: 39347998
-
Comparative genomic analysis of immune-related genes and chemosensory receptors provides insights into the evolution and adaptation of four major domesticated Asian carps.BMC Genomics. 2025 May 26;26(1):529. doi: 10.1186/s12864-025-11719-2. BMC Genomics. 2025. PMID: 40419972 Free PMC article.
-
The spatiotemporal and genetic architecture of extraoral taste buds in Astyanax cavefish.Commun Biol. 2024 Aug 6;7(1):951. doi: 10.1038/s42003-024-06635-2. Commun Biol. 2024. PMID: 39107459 Free PMC article.
-
Genomic signatures of sensory adaptation and evolution in pangolins.BMC Genomics. 2024 Dec 4;25(1):1176. doi: 10.1186/s12864-024-11063-x. BMC Genomics. 2024. PMID: 39633301 Free PMC article.
-
Gene networks and the evolution of olfactory organs, eyes, hair cells and motoneurons: a view encompassing lancelets, tunicates and vertebrates.Front Cell Dev Biol. 2024 Mar 12;12:1340157. doi: 10.3389/fcell.2024.1340157. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38533086 Free PMC article. Review.
References
-
- Müller J, Bickelmann C, Sobral G. The Evolution and Fossil History of Sensory Perception in Amniote Vertebrates. Annu. Rev. Earth Planet. Sci. 2018;46:495–519. doi: 10.1146/annurev-earth-082517-010120. - DOI
-
- Stevens, M. Sensory Ecology, Behaviour, and Evolution (Oxford University Press, 2013).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

