High resolution spatial profiling of kidney injury and repair using RNA hybridization-based in situ sequencing
- PMID: 38360882
- PMCID: PMC10869771
- DOI: 10.1038/s41467-024-45752-8
High resolution spatial profiling of kidney injury and repair using RNA hybridization-based in situ sequencing
Abstract
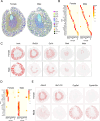
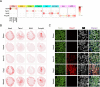
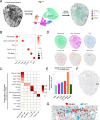
Emerging spatially resolved transcriptomics technologies allow for the measurement of gene expression in situ at cellular resolution. We apply direct RNA hybridization-based in situ sequencing (dRNA HybISS, Cartana part of 10xGenomics) to compare male and female healthy mouse kidneys and the male kidney injury and repair timecourse. A pre-selected panel of 200 genes is used to identify cell state dynamics patterns during injury and repair. We develop a new computational pipeline, CellScopes, for the rapid analysis, multi-omic integration and visualization of spatially resolved transcriptomic datasets. The resulting dataset allows us to resolve 13 kidney cell types within distinct kidney niches, dynamic alterations in cell state over the course of injury and repair and cell-cell interactions between leukocytes and kidney parenchyma. At late timepoints after injury, C3+ leukocytes are enriched near pro-inflammatory, failed-repair proximal tubule cells. Integration of snRNA-seq dataset from the same injury and repair samples also allows us to impute the spatial localization of genes not directly measured by dRNA HybISS.
© 2024. The Author(s).
Conflict of interest statement
B.D.H. is a consultant for Janssen Research & Development, LLC, Pfizer, Roche and Chinook Therapeutics, holds equity in Chinook Therapeutics and grant funding from Pfizer, all unrelated to the current study. D.C., H.X., A.N. and M.R. were employees of 10X Genomics during this study. M.R. holds stock options in 10X Genomics. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

