The Drosophila ZNRF1/2 homologue, detour, interacts with HOPS complex and regulates autophagy
- PMID: 38360932
- PMCID: PMC10869362
- DOI: 10.1038/s42003-024-05834-1
The Drosophila ZNRF1/2 homologue, detour, interacts with HOPS complex and regulates autophagy
Abstract
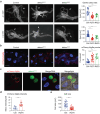
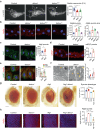
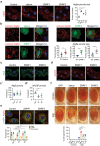
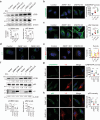
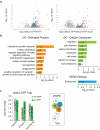
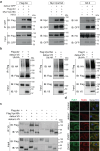
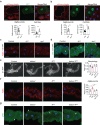
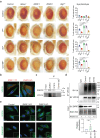
Autophagy, the process of elimination of cellular components by lysosomal degradation, is essential for animal development and homeostasis. Using the autophagy-dependent Drosophila larval midgut degradation model we identified an autophagy regulator, the RING domain ubiquitin ligase CG14435 (detour). Depletion of detour resulted in increased early-stage autophagic vesicles, premature tissue contraction, and overexpression of detour or mammalian homologues, ZNRF1 and ZNRF2, increased autophagic vesicle size. The ablation of ZNRF1 or ZNRF2 in mammalian cells increased basal autophagy. We identified detour interacting proteins including HOPS subunits, deep orange (dor/VPS18), Vacuolar protein sorting 16A (VPS16A), and light (lt/VPS41) and found that detour promotes their ubiquitination. The detour mutant accumulated autophagy-related proteins in young adults, displayed premature ageing, impaired motor function, and activation of innate immunity. Collectively, our findings suggest a role for detour in autophagy, likely through regulation of HOPS complex, with implications for healthy aging.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest regarding the publication of this paper.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

