Modular, multi-robot integration of laboratories: an autonomous workflow for solid-state chemistry
- PMID: 38362408
- PMCID: PMC10866346
- DOI: 10.1039/d3sc06206f
Modular, multi-robot integration of laboratories: an autonomous workflow for solid-state chemistry
Abstract
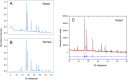
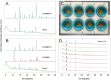
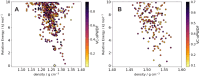
Automation can transform productivity in research activities that use liquid handling, such as organic synthesis, but it has made less impact in materials laboratories, which require sample preparation steps and a range of solid-state characterization techniques. For example, powder X-ray diffraction (PXRD) is a key method in materials and pharmaceutical chemistry, but its end-to-end automation is challenging because it involves solid powder handling and sample processing. Here we present a fully autonomous solid-state workflow for PXRD experiments that can match or even surpass manual data quality, encompassing crystal growth, sample preparation, and automated data capture. The workflow involves 12 steps performed by a team of three multipurpose robots, illustrating the power of flexible, modular automation to integrate complex, multitask laboratories.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources

