Cytotoxic Programming of CD4+ T Cells Is Regulated by Opposing Actions of the Related Transcription Factors Eos and Aiolos
- PMID: 38363226
- PMCID: PMC10948294
- DOI: 10.4049/jimmunol.2300748
Cytotoxic Programming of CD4+ T Cells Is Regulated by Opposing Actions of the Related Transcription Factors Eos and Aiolos
Abstract
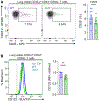
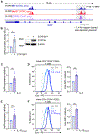
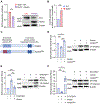
In contrast to the "helper" activities of most CD4+ T effector subsets, CD4+ cytotoxic T lymphocytes (CD4-CTLs) perform functions normally associated with CD8+ T and NK cells. Specifically, CD4-CTLs secrete cytotoxic molecules and directly target and kill compromised cells in an MHC class II-restricted fashion. The functions of these cells have been described in diverse immunological contexts, including their ability to provide protection during antiviral and antitumor responses, as well as being implicated in autoimmunity. Despite their significance to human health, the complete mechanisms that govern their programming remain unclear. In this article, we identify the Ikaros zinc finger transcription factor Eos (Ikzf4) as a positive regulator of CD4-CTL differentiation during murine immune responses against influenza virus infection. We find that the frequency of Eos+ cells is elevated in lung CD4-CTL populations and that the cytotoxic gene program is compromised in Eos-deficient CD4+ T cells. Consequently, we observe a reduced frequency and number of lung-residing, influenza virus-responsive CD4-CTLs in the absence of Eos. Mechanistically, we determine that this is due, at least in part, to reduced expression of IL-2 and IL-15 cytokine receptor subunits on the surface of Eos-deficient CD4+ T cells, both of which support the CD4-CTL program. Finally, we find that Aiolos, a related Ikaros family member and known CD4-CTL antagonist, represses Eos expression by antagonizing STAT5-dependent activation of the Ikzf4 promoter. Collectively, our findings reveal a mechanism wherein Eos and Aiolos act in opposition to regulate cytotoxic programming of CD4+ T cells.
Copyright © 2024 by The American Association of Immunologists, Inc.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

