Antigenic strain diversity predicts different biogeographic patterns of maintenance and decline of antimalarial drug resistance
- PMID: 38363295
- PMCID: PMC10942604
- DOI: 10.7554/eLife.90888
Antigenic strain diversity predicts different biogeographic patterns of maintenance and decline of antimalarial drug resistance
Abstract
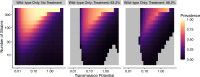
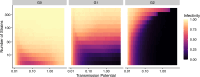
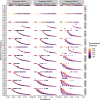
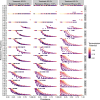
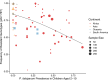
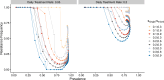
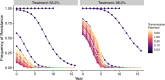
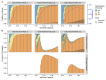
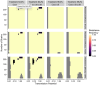
The establishment and spread of antimalarial drug resistance vary drastically across different biogeographic regions. Though most infections occur in sub-Saharan Africa, resistant strains often emerge in low-transmission regions. Existing models on resistance evolution lack consensus on the relationship between transmission intensity and drug resistance, possibly due to overlooking the feedback between antigenic diversity, host immunity, and selection for resistance. To address this, we developed a novel compartmental model that tracks sensitive and resistant parasite strains, as well as the host dynamics of generalized and antigen-specific immunity. Our results show a negative correlation between parasite prevalence and resistance frequency, regardless of resistance cost or efficacy. Validation using chloroquine-resistant marker data supports this trend. Post discontinuation of drugs, resistance remains high in low-diversity, low-transmission regions, while it steadily decreases in high-diversity, high-transmission regions. Our study underscores the critical role of malaria strain diversity in the biogeographic patterns of resistance evolution.
Keywords: Greater Mekong Subregion; P. falciparum; antigenic diversity; disease modeling; drug resistance; epidemiology; global health.
Plain language summary
Drug resistance among strains of the parasites that cause malaria is a growing problem for people relying on antimalarial drugs to protect them from the disease. This phenomenon is global yet exactly how resistance emerges, spreads and persists in a population often differs greatly between regions, which can complicate malaria control projects. For example, discontinuing the use of antimalarials can lead to the frequency of resistant strains declining in an area, such as Africa, but persisting at high levels in others, including Asia and South America. Gaining resistance often leads to parasites becoming less transmissible than other strains. When antimalarials are not used, sensitive strains usually outcompete their resistant counterparts. However, prolonged use of antimalarial drugs tends to eliminate susceptible strains, allowing the previously outcompeted resistant strains to dominate. The local dynamics of antimalarial resistance are also shaped by multiple other factors such as transmission levels (how common the disease is in the region), the type of antimalarial measures used (such as drugs and mosquito nets), or previous immunity the population may have developed to specific strains. While many computational models have been developed to capture these dynamics, they usually fail to include strain diversity – a parameter reflecting the number of malaria strains the immune system is exposed to. This parameter is important as parasites need to escape both host immunity and drugs in order to be successful. To address this gap, He, Chaillet, and Labbé created a computational model to investigate how strain diversity, transmission levels and other related factors influence antimalarial resistance. The model was used to explore how the frequency of resistant and susceptible strains changes over time once antimalarial drugs are rolled out and then halted. These analyses show that in areas with both low strain diversity and low transmission levels, susceptible parasites are more likely to be wiped out from the population, leading to a high frequency of resistant strains that persist after drugs are discontinued. However, in high diversity and high transmission regions, susceptible strains can remain in the population. Therefore, when drug treatments are stopped, resistance levels are more likely to drop due to these parasites outcompeting the drug-resistant ones. Overall, this work demonstrates how modelling approaches that include strain diversity can help inform public health decisions aimed at reducing antimalarial resistance. In particular, they can provide important insights into the control strategies that are best suited for a specific region, suggesting that in low transmission areas intensive drug treatment may contribute to resistance. Instead, preventative strategies such as eliminating mosquitos and preventing bites with bed nets may prove more beneficial at reducing transmission rates in such areas.
© 2023, He, Chaillet et al.
Conflict of interest statement
QH, JC, FL No competing interests declared
Figures

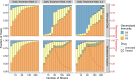
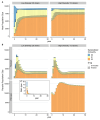
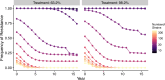
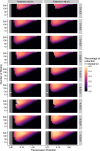

Update of
-
Antigenic strain diversity predicts different biogeographic patterns of maintenance and decline of anti-malarial drug resistance.bioRxiv [Preprint]. 2023 Dec 5:2023.03.06.531320. doi: 10.1101/2023.03.06.531320. bioRxiv. 2023. Update in: Elife. 2024 Feb 16;12:RP90888. doi: 10.7554/eLife.90888. PMID: 37987011 Free PMC article. Updated. Preprint.
Similar articles
-
Antigenic strain diversity predicts different biogeographic patterns of maintenance and decline of anti-malarial drug resistance.bioRxiv [Preprint]. 2023 Dec 5:2023.03.06.531320. doi: 10.1101/2023.03.06.531320. bioRxiv. 2023. Update in: Elife. 2024 Feb 16;12:RP90888. doi: 10.7554/eLife.90888. PMID: 37987011 Free PMC article. Updated. Preprint.
-
The Rare, the Best: Spread of Antimalarial-Resistant Plasmodium falciparum Parasites by Anopheles Mosquito Vectors.Microbiol Spectr. 2021 Oct 31;9(2):e0085221. doi: 10.1128/Spectrum.00852-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668767 Free PMC article.
-
Within-host competition can delay evolution of drug resistance in malaria.PLoS Biol. 2018 Aug 21;16(8):e2005712. doi: 10.1371/journal.pbio.2005712. eCollection 2018 Aug. PLoS Biol. 2018. PMID: 30130363 Free PMC article.
-
The mechanisms of resistance to antimalarial drugs in Plasmodium falciparum.Fundam Clin Pharmacol. 2003 Apr;17(2):147-53. doi: 10.1046/j.1472-8206.2003.00164.x. Fundam Clin Pharmacol. 2003. PMID: 12667224 Review.
-
Population biology and antimalarial resistance: The transmission of antimalarial drug resistance in Plasmodium falciparum.Acta Trop. 2005 Jun;94(3):230-40. doi: 10.1016/j.actatropica.2005.04.014. Acta Trop. 2005. PMID: 15878154 Review.
Cited by
-
Artemisinin and partner drug resistance markers in Plasmodium falciparum from Tanzanian paediatric malaria patients, 2016-2022.Malar J. 2025 Jul 1;24(1):209. doi: 10.1186/s12936-025-05447-x. Malar J. 2025. PMID: 40598187 Free PMC article.
References
-
- Albrecht L, Castiñeiras C, Carvalho BO, Ladeia-Andrade S, Santos da Silva N, Hoffmann EHE, dalla Martha RC, Costa FTM, Wunderlich G. The South American Plasmodium falciparum var gene repertoire is limited, highly shared and possibly lacks several antigenic types. Gene. 2010;453:37–44. doi: 10.1016/j.gene.2010.01.001. - DOI - PubMed
-
- Alifrangis M, Lemnge MM, Rønn AM, Segeja MD, Magesa SM, Khalil IF, Bygbjerg IC. Increasing prevalence of wildtypes in the dihydrofolate reductase gene of Plasmodium falciparum in an area with high levels of sulfadoxine/pyrimethamine resistance after introduction of treated bed nets. The American Journal of Tropical Medicine and Hygiene. 2003;69:238–243. - PubMed
-
- Anderson RM, May RM. REgulation and stability of host-parasite population interactions: I. Regulatory processes. The Journal of Animal Ecology. 1978;47:219. doi: 10.2307/3933. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

