A Novel Heterozygous Variant in AICDA Impairs Ig Class Switching and Somatic Hypermutation in Human B Cells and is Associated with Autosomal Dominant HIGM2 Syndrome
- PMID: 38363477
- PMCID: PMC10873450
- DOI: 10.1007/s10875-024-01665-1
A Novel Heterozygous Variant in AICDA Impairs Ig Class Switching and Somatic Hypermutation in Human B Cells and is Associated with Autosomal Dominant HIGM2 Syndrome
Abstract
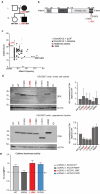
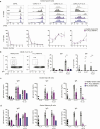
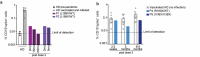
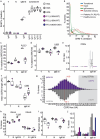
B cells and their secreted antibodies are fundamental for host-defense against pathogens. The generation of high-affinity class switched antibodies results from both somatic hypermutation (SHM) of the immunoglobulin (Ig) variable region genes of the B-cell receptor and class switch recombination (CSR) which alters the Ig heavy chain constant region. Both of these processes are initiated by the enzyme activation-induced cytidine deaminase (AID), encoded by AICDA. Deleterious variants in AICDA are causal of hyper-IgM syndrome type 2 (HIGM2), a B-cell intrinsic primary immunodeficiency characterised by recurrent infections and low serum IgG and IgA levels. Biallelic variants affecting exons 2, 3 or 4 of AICDA have been identified that impair both CSR and SHM in patients with autosomal recessive HIGM2. Interestingly, B cells from patients with autosomal dominant HIGM2, caused by heterozygous variants (V186X, R190X) located in AICDA exon 5 encoding the nuclear export signal (NES) domain, show abolished CSR but variable SHM. We herein report the immunological and functional phenotype of two related patients presenting with common variable immunodeficiency who were found to have a novel heterozygous variant in AICDA (L189X). This variant led to a truncated AID protein lacking the last 10 amino acids of the NES at the C-terminal domain. Interestingly, patients' B cells carrying the L189X variant exhibited not only greatly impaired CSR but also SHM in vivo, as well as CSR and production of IgG and IgA in vitro. Our findings demonstrate that the NES domain of AID can be essential for SHM, as well as for CSR, thereby refining the correlation between AICDA genotype and SHM phenotype as well as broadening our understanding of the pathophysiology of HIGM disorders.
Keywords: AICDA; HIGM2; Human B cells; Ig class switching; Somatic hypermutation.
© 2024. The Author(s).
Conflict of interest statement
All authors have no conflicts of interest to declare.
Figures

Similar articles
-
A Novel AICDA Splice-Site Mutation in Two Siblings with HIGM2 Permits Somatic Hypermutation but Abrogates Mutational Targeting.J Clin Immunol. 2022 May;42(4):771-782. doi: 10.1007/s10875-022-01233-5. Epub 2022 Mar 5. J Clin Immunol. 2022. PMID: 35246784 Free PMC article.
-
Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2.Clin Immunol. 2005 Jun;115(3):277-85. doi: 10.1016/j.clim.2005.02.003. Clin Immunol. 2005. PMID: 15893695
-
A novel activation-induced cytidine deaminase (AID) mutation in Brazilian patients with hyper-IgM type 2 syndrome.Clin Immunol. 2013 Aug;148(2):279-86. doi: 10.1016/j.clim.2013.05.017. Epub 2013 Jun 7. Clin Immunol. 2013. PMID: 23803409
-
Regulation of aicda expression and AID activity: relevance to somatic hypermutation and class switch DNA recombination.Crit Rev Immunol. 2007;27(4):367-97. doi: 10.1615/critrevimmunol.v27.i4.60. Crit Rev Immunol. 2007. PMID: 18197815 Free PMC article. Review.
-
Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies.Adv Immunol. 2007;94:275-306. doi: 10.1016/S0065-2776(06)94009-7. Adv Immunol. 2007. PMID: 17560278 Review.
Cited by
-
Faecalibacterium prausnitzii enhances intestinal IgA response by host-microbe derived inecalcitol in colitis.BMC Med. 2025 Jul 15;23(1):425. doi: 10.1186/s12916-025-04260-2. BMC Med. 2025. PMID: 40660288 Free PMC article.
References
-
- Banchereau J, Rousset F. Human B lymphocytes: phenotype, proliferation, and differentiation. Adv Immunol. 1992;52:125–262. - PubMed
-
- Liu YJ, de Bouteiller O, Fugier-Vivier I. Mechanisms of selection and differentiation in germinal centers. Curr Opin Immunol. 1997;9(2):256–262. - PubMed
-
- Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11(8):681–688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

