Genomics of the expanding pine pathogen Lecanosticta acicola reveals patterns of ongoing genetic admixture
- PMID: 38364101
- PMCID: PMC10949461
- DOI: 10.1128/msystems.00928-23
Genomics of the expanding pine pathogen Lecanosticta acicola reveals patterns of ongoing genetic admixture
Abstract
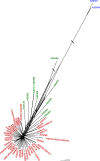
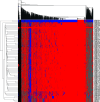
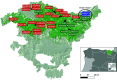
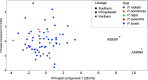
Lecanosticta acicola is the causal agent for brown spot needle blight that affects pine trees across the northern hemisphere. Based on marker genes and microsatellite data, two distinct lineages have been identified that were introduced into Europe on two separate occasions. Despite their overall distinct geographic distribution, they have been found to coexist in regions of northern Spain and France. Here, we present the first genome-wide study of Lecanosticta acicola, including assembly of the reference genome and a population genomics analysis of 70 natural isolates from northern Spain. We show that most of the isolates belong to the southern lineage but show signs of introgression with northern lineage isolates, indicating mating between the two lineages. We also identify phenotypic differences between the two lineages based on the activity profiles of 20 enzymes, with introgressed strains being more phenotypically similar to members of the southern lineage. In conclusion, we show undergoing genetic admixture between the two main lineages of L. acicola in a region of recent expansion.
Importance: Lecanosticta acicola is a fungal pathogen causing severe defoliation, growth reduction, and even death in more than 70 conifer species. Despite the increasing incidence of this species, little is known about its population dynamics. Two divergent lineages have been described that have now been found together in regions of France and Spain, but it is unknown how these mixed populations evolve. Here we present the first reference genome for this important plant pathogenic fungi and use it to study the population genomics of 70 isolates from an affected forest in the north of Spain. We find signs of introgression between the two main lineages, indicating that active mating is occurring in this region which could propitiate the appearance of novel traits in this species. We also study the phenotypic differences across this population based on enzymatic activities on 20 compounds.
Keywords: Lecanosticta acicola; admixture; comparative genomics; needle blight; plant pathogen; population genomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Remote sensing-based detection of brown spot needle blight: a comprehensive review, and future directions.PeerJ. 2025 May 22;13:e19407. doi: 10.7717/peerj.19407. eCollection 2025. PeerJ. 2025. PMID: 40416626 Free PMC article. Review.
-
The increasing threat to European forests from the invasive foliar pine pathogen, Lecanosticta acicola.For Ecol Manage. 2023 May 15;536:120847. doi: 10.1016/j.foreco.2023.120847. For Ecol Manage. 2023. PMID: 37193248 Free PMC article.
-
Lecanosticta acicola: A growing threat to expanding global pine forests and plantations.Mol Plant Pathol. 2019 Oct;20(10):1327-1364. doi: 10.1111/mpp.12853. Epub 2019 Jul 15. Mol Plant Pathol. 2019. PMID: 31309681 Free PMC article.
-
Diversity, migration routes, and worldwide population genetic structure of Lecanosticta acicola, the causal agent of brown spot needle blight.Mol Plant Pathol. 2022 Nov;23(11):1620-1639. doi: 10.1111/mpp.13257. Epub 2022 Aug 11. Mol Plant Pathol. 2022. PMID: 35957598 Free PMC article.
-
Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics.Int J Med Microbiol. 2011 Feb;301(2):79-96. doi: 10.1016/j.ijmm.2010.05.002. Epub 2010 Aug 13. Int J Med Microbiol. 2011. PMID: 20708964 Review.
Cited by
-
Remote sensing-based detection of brown spot needle blight: a comprehensive review, and future directions.PeerJ. 2025 May 22;13:e19407. doi: 10.7717/peerj.19407. eCollection 2025. PeerJ. 2025. PMID: 40416626 Free PMC article. Review.
References
-
- Mesanza N, García-García D, Raposo ER, Raposo R, Iturbide M, Pascual MT, Barrena I, Urkola A, Berano N, Sáez de Zerain A, Iturritxa E. 2021. Weather variables associated with spore dispersal of Lecanosticta acicola causing pine needle blight in northern Spain. Plants 10:2788. doi:10.3390/plants10122788 - DOI - PMC - PubMed
-
- Tubby K, Adamčikova K, Adamson K, Akiba M, Barnes I, Boroń P, Bragança H, Bulgakov T, Burgdorf N, Capretti P, et al. . 2023. The increasing threat to European forests from the invasive foliar pine pathogen, Lecanosticta acicola. For Ecol Manage 536:120847. doi:10.1016/j.foreco.2023.120847 - DOI - PMC - PubMed
-
- Ortíz de Urbina E, Mesanza N, Aragonés A, Raposo R, Elvira-Recuenco M, Boqué R, Patten C, Aitken J, Iturritxa E. 2016. Emerging needle blight diseases in Atlantic pinus ecosystems of Spain. For Trees Livelihood 8:18. doi:10.3390/f8010018 - DOI
-
- Aglietti C, Meinecke CD, Ghelardini L, Barnes I, van der Nest A, Villari C. 2021. Rapid detection of pine pathogens Lecanosticta acicola, Dothistroma pini and D. septosporum on needles by probe-based LAMP assays. For Trees Livelihoods 12:479. doi:10.3390/f12040479 - DOI
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

