Akkermansia muciniphila-induced trained immune phenotype increases bacterial intracellular survival and attenuates inflammation
- PMID: 38365881
- PMCID: PMC10873422
- DOI: 10.1038/s42003-024-05867-6
Akkermansia muciniphila-induced trained immune phenotype increases bacterial intracellular survival and attenuates inflammation
Abstract
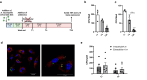
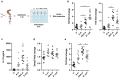
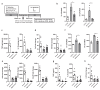
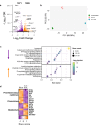
The initial exposure to pathogens and commensals confers innate immune cells the capacity to respond distinctively upon a second stimulus. This training capacity might play key functions in developing an adequate innate immune response to the continuous exposure to bacteria. However, the mechanisms involved in induction of trained immunity by commensals remain mostly unexplored. A. muciniphila represents an attractive candidate to study the promotion of these long-term responses. Here, we show that priming of macrophages with live A. muciniphila enhances bacterial intracellular survival and decreases the release of pro- and anti-inflammatory signals, lowering the production of TNF and IL-10. Global transcriptional analysis of macrophages after a secondary exposure to the bacteria showed the transcriptional rearrangement underpinning the phenotype observed compared to acutely exposed cells, with the increased expression of genes related to phagocytic capacity and those involved in the metabolic adjustment conducing to innate immune training. Accordingly, key genes related to bacterial killing and pro-inflammatory pathways were downregulated. These data demonstrate the importance of specific bacterial members in the modulation of local long-term innate immune responses, broadening our knowledge of the association between gut microbiome commensals and trained immunity as well as the anti-inflammatory probiotic potential of A. muciniphila.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

