A divergent intermediate strategy yields biologically diverse pseudo-natural products
- PMID: 38365941
- PMCID: PMC11164679
- DOI: 10.1038/s41557-024-01458-4
A divergent intermediate strategy yields biologically diverse pseudo-natural products
Abstract
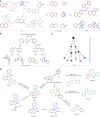
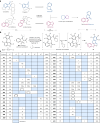
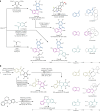
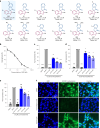
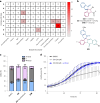
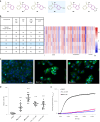
The efficient exploration of biologically relevant chemical space is essential for the discovery of bioactive compounds. A molecular design principle that possesses both biological relevance and structural diversity may more efficiently lead to compound collections that are enriched in diverse bioactivities. Here the diverse pseudo-natural product (PNP) strategy, which combines the biological relevance of the PNP concept with synthetic diversification strategies from diversity-oriented synthesis, is reported. A diverse PNP collection was synthesized from a common divergent intermediate through developed indole dearomatization methodologies to afford three-dimensional molecular frameworks that could be further diversified via intramolecular coupling and/or carbon monoxide insertion. In total, 154 PNPs were synthesized representing eight different classes. Cheminformatic analyses showed that the PNPs are structurally diverse between classes. Biological investigations revealed the extent of diverse bioactivity enrichment of the collection in which four inhibitors of Hedgehog signalling, DNA synthesis, de novo pyrimidine biosynthesis and tubulin polymerization were identified from four different PNP classes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

