Microbial eukaryotic predation pressure and biomass at deep-sea hydrothermal vents
- PMID: 38366040
- PMCID: PMC10939315
- DOI: 10.1093/ismejo/wrae004
Microbial eukaryotic predation pressure and biomass at deep-sea hydrothermal vents
Erratum in
-
Correction to 29 articles due to inaccurate manuscript submission dates.ISME J. 2025 Jan 2;19(1):wraf008. doi: 10.1093/ismejo/wraf008. ISME J. 2025. PMID: 39981677 Free PMC article. No abstract available.
Abstract
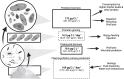
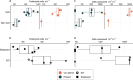
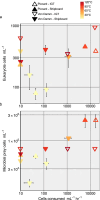
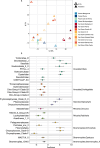
Deep-sea hydrothermal vent geochemistry shapes the foundation of the microbial food web by fueling chemolithoautotrophic microbial activity. Microbial eukaryotes (or protists) play a critical role in hydrothermal vent food webs as consumers and hosts of symbiotic bacteria, and as a nutritional source to higher trophic levels. We measured microbial eukaryotic cell abundance and predation pressure in low-temperature diffuse hydrothermal fluids at the Von Damm and Piccard vent fields along the Mid-Cayman Rise in the Western Caribbean Sea. We present findings from experiments performed under in situ pressure that show cell abundances and grazing rates higher than those done at 1 atmosphere (shipboard ambient pressure); this trend was attributed to the impact of depressurization on cell integrity. A relationship between the protistan grazing rate, prey cell abundance, and temperature of end-member hydrothermal vent fluid was observed at both vent fields, regardless of experimental approach. Our results show substantial protistan biomass at hydrothermally fueled microbial food webs, and when coupled with improved grazing estimates, suggest an important contribution of grazers to the local carbon export and supply of nutrient resources to the deep ocean.
Keywords: deep-sea food web ecology; deep-sea hydrothermal vents; microbial eukaryotes; predator–prey interactions; protists.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
Authors declare no competing interests.
Figures
References
-
- Van Dover CL. The Ecology of Deep-Sea Hydrothermal Vents, 1st edn. Princeton: Princeton University Press, 2000
-
- Tunnicliffe V, McArthur AG, McHugh D. A biogeographical perspective of the deep-sea hydrothermal vent fauna. In: Blaxter J.H.S., Southward A.J., Tyler P.A. (eds.), Advances in Marine Biology. New York: Academic Press, 1998, 353–442
MeSH terms
Grants and funding
- OCE-1947776/National Science Foundation
- Center for Dark Energy Biosphere Investigations
- OCE-0939564/C-DEBI Postdoctoral Fellowship
- Charles E. Hollister Endowed Fund for Support of Innovative Research at Woods Hole Oceanographic Institution
- 80NSSC17K0252/NASA Planetary Science and Technology Through Analog Research
LinkOut - more resources
Full Text Sources

