Coral mucus as a reservoir of bacteriophages targeting Vibrio pathogens
- PMID: 38366190
- PMCID: PMC10945359
- DOI: 10.1093/ismejo/wrae017
Coral mucus as a reservoir of bacteriophages targeting Vibrio pathogens
Abstract
The increasing trend in sea surface temperature promotes the spread of Vibrio species, which are known to cause diseases in a wide range of marine organisms. Among these pathogens, Vibrio mediterranei has emerged as a significant threat, leading to bleaching in the coral species Oculina patagonica. Bacteriophages, or phages, are viruses that infect bacteria, thereby regulating microbial communities and playing a crucial role in the coral's defense against pathogens. However, our understanding of phages that infect V. mediterranei is limited. In this study, we identified two phage species capable of infecting V. mediterranei by utilizing a combination of cultivation and metagenomic approaches. These phages are low-abundance specialists within the coral mucus layer that exhibit rapid proliferation in the presence of their hosts, suggesting a potential role in coral defense. Additionally, one of these phages possesses a conserved domain of a leucine-rich repeat protein, similar to those harbored in the coral genome, that plays a key role in pathogen recognition, hinting at potential coral-phage coevolution. Furthermore, our research suggests that lytic Vibrio infections could trigger prophage induction, which may disseminate genetic elements, including virulence factors, in the coral mucus layer. Overall, our findings underscore the importance of historical coral-phage interactions as a form of coral immunity against invasive Vibrio pathogens.
Keywords: Vibrio mediterranei; coral mucus layer; vibriophage.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures

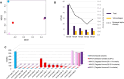

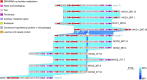

Similar articles
-
Virulence as a Side Effect of Interspecies Interaction in Vibrio Coral Pathogens.mBio. 2020 Jul 21;11(4):e00201-20. doi: 10.1128/mBio.00201-20. mBio. 2020. PMID: 32694137 Free PMC article.
-
Structure and temporal dynamics of the bacterial communities associated to microhabitats of the coral Oculina patagonica.Environ Microbiol. 2016 Dec;18(12):4564-4578. doi: 10.1111/1462-2920.13548. Environ Microbiol. 2016. PMID: 27690185
-
Changes in the diversity and functionality of viruses that can bleach healthy coral.mSphere. 2024 Dec 19;9(12):e0081624. doi: 10.1128/msphere.00816-24. Epub 2024 Nov 26. mSphere. 2024. PMID: 39589125 Free PMC article.
-
The Vibrio shiloi/Oculina patagonica model system of coral bleaching.Annu Rev Microbiol. 2004;58:143-59. doi: 10.1146/annurev.micro.58.030603.123610. Annu Rev Microbiol. 2004. PMID: 15487933 Review.
-
The Role of Vibrios in Diseases of Corals.Microbiol Spectr. 2015 Aug;3(4). doi: 10.1128/microbiolspec.VE-0006-2014. Microbiol Spectr. 2015. PMID: 26350314 Review.
Cited by
-
Ecophysiological responses to heat waves in the marine intertidal zone.J Exp Biol. 2025 Jan 15;228(2):JEB246503. doi: 10.1242/jeb.246503. Epub 2025 Jan 16. J Exp Biol. 2025. PMID: 39817480 Free PMC article. Review.
-
Recent Discovery of Diverse Prophages Located in Genomes of Vibrio spp. and Their Implications for Bacterial Pathogenicity, Environmental Fitness, Genome Evolution, Food Safety, and Public Health.Foods. 2025 Jan 26;14(3):403. doi: 10.3390/foods14030403. Foods. 2025. PMID: 39941999 Free PMC article. Review.
References
-
- Vega Thurber R, Mydlarz LD, Brandt Met al. . Deciphering coral disease dynamics: integrating host microbiome and the changing environment. Front Ecol Environ 2020;2020:575927.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

