Tumor acidosis-induced DNA damage response and tetraploidy enhance sensitivity to ATM and ATR inhibitors
- PMID: 38366255
- PMCID: PMC10933359
- DOI: 10.1038/s44319-024-00089-7
Tumor acidosis-induced DNA damage response and tetraploidy enhance sensitivity to ATM and ATR inhibitors
Abstract
Tumor acidosis is associated with increased invasiveness and drug resistance. Here, we take an unbiased approach to identify vulnerabilities of acid-exposed cancer cells by combining pH-dependent flow cytometry cell sorting from 3D colorectal tumor spheroids and transcriptomic profiling. Besides metabolic rewiring, we identify an increase in tetraploid cell frequency and DNA damage response as consistent hallmarks of acid-exposed cancer cells, supported by the activation of ATM and ATR signaling pathways. We find that regardless of the cell replication error status, both ATM and ATR inhibitors exert preferential growth inhibitory effects on acid-exposed cancer cells. The efficacy of a combination of these drugs with 5-FU is further documented in 3D spheroids as well as in patient-derived colorectal tumor organoids. These data position tumor acidosis as a revelator of the therapeutic potential of DNA repair blockers and as an attractive clinical biomarker to predict the response to a combination with chemotherapy.
Keywords: 3D Spheroids; ATM; DNA Damage Response; Organoids; Tumor Acidosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



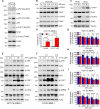



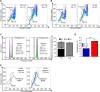

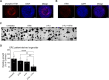
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

