Phenotypic and genetic characterization of antimicrobial resistance in Salmonella enterica serovar Choleraesuis isolates from humans and animals in Spain from 2006 to 2021
- PMID: 38366818
- PMCID: PMC10984937
- DOI: 10.1093/jac/dkae029
Phenotypic and genetic characterization of antimicrobial resistance in Salmonella enterica serovar Choleraesuis isolates from humans and animals in Spain from 2006 to 2021
Abstract
Objectives: While an increase in the levels of MDR in Salmonella enterica sevorar Choleraesuis has been reported in Europe, little is known about the situation in Spain. Therefore, we first aimed to assess the phenotypic resistance profile and to determine the presence of genetic determinants of resistance of S. Choleraesuis isolates collected in animal and human. Our second objective was to identify and characterize clusters of highly related isolates.
Methods: We analysed 50 human and 45 animal isolates retrieved from 2006 to 2021 using the disc diffusion method and performed WGS followed by analyses of genetic determinants and phylogenetic analysis.
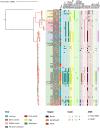
Results: All isolates were of ST145 and corresponded to the variant Kunzendorf. Swine isolates harboured a significantly higher number of antimicrobial resistance genes than human isolates, and often carried plasmid replicons of the IncHI2/IncHI2A type (42% of all animal isolates). In addition, we identified several MDR S. Choleraesuis strains circulating in humans and swine between 2006 and 2021. The phylogenetic analyses identified four clades associated with specific patterns of resistance genes and plasmid replicons. The clades also included isolates that differed in terms of year and region of isolation as well as host of origin.
Conclusions: This One Health approach highlights that reducing human MDR S. Choleraesuis infections may require the adoption of strategies that not only seek to prevent cases in humans but also to characterize and reduce the infection burden in swine.
© The Author(s) 2024. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures



Similar articles
-
Outbreaks of antimicrobial resistant Salmonella Choleraesuis in wild boars piglets from central-western Spain.Transbound Emerg Dis. 2019 Jan;66(1):225-233. doi: 10.1111/tbed.13003. Epub 2018 Sep 10. Transbound Emerg Dis. 2019. PMID: 30144295 Free PMC article.
-
Antimicrobial resistance in Salmonella enterica serovar Choleraesuis isolated from pigs in northern Italy.Res Vet Sci. 2025 Sep;193:105773. doi: 10.1016/j.rvsc.2025.105773. Epub 2025 Jun 17. Res Vet Sci. 2025. PMID: 40570720
-
Antimicrobial resistance profile in Salmonella enterica serovar Choleraesuis isolates from diseased pigs in Taiwan.J Microbiol Immunol Infect. 2024 Aug;57(4):660-664. doi: 10.1016/j.jmii.2024.04.005. Epub 2024 Apr 16. J Microbiol Immunol Infect. 2024. PMID: 38670815
-
Genomic characterization of Salmonella enterica serovar Choleraesuis from Brazil reveals a swine gallbladder isolate harboring colistin resistance gene mcr-1.1.Braz J Microbiol. 2022 Dec;53(4):1799-1806. doi: 10.1007/s42770-022-00812-3. Epub 2022 Aug 19. Braz J Microbiol. 2022. PMID: 35984599 Free PMC article.
-
Molecular typing and antimicrobial resistance of Salmonella enterica subspecies enterica serovar Choleraesuis isolates from diseased pigs in Japan.Comp Immunol Microbiol Infect Dis. 2010 Mar;33(2):109-19. doi: 10.1016/j.cimid.2008.08.004. Epub 2008 Sep 21. Comp Immunol Microbiol Infect Dis. 2010. PMID: 18809209
References
-
- Reed WM, Olander HJ, Thacker HL. Studies on the pathogenesis of Salmonella typhimurium and Salmonella choleraesuis var kunzendorf infection in weanling pigs. Am J Vet Res 1986; 47: 75–83. - PubMed

