A simple, robust, broadly applicable insertion mutagenesis method to create random fluorescent protein: target protein fusions
- PMID: 38366837
- PMCID: PMC11075570
- DOI: 10.1093/g3journal/jkae036
A simple, robust, broadly applicable insertion mutagenesis method to create random fluorescent protein: target protein fusions
Abstract
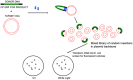
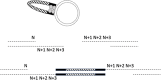
A simple, broadly applicable method was developed using an in vitro transposition reaction followed by transformation into Escherichia coli and screening plates for fluorescent colonies. The transposition reaction catalyzes the random insertion of a fluorescent protein open reading frame into a target gene on a plasmid. The transposition reaction is employed directly in an E. coli transformation with no further procedures. Plating at high colony density yields fluorescent colonies. Plasmids purified from fluorescent colonies contain random, in-frame fusion proteins into the target gene. The plate screen also results in expressed, stable proteins. A large library of chimeric proteins was produced, which was useful for downstream research. The effect of using different fluorescent proteins was investigated as well as the dependence of the linker sequence between the target and fluorescent protein open reading frames. The utility and simplicity of the method were demonstrated by the fact that it has been employed in an undergraduate biology laboratory class without failure over dozens of class sections. This suggests that the method will be useful in high-impact research at small liberal arts colleges with limited resources. However, in-frame fusion proteins were obtained from 8 different targets suggesting that the method is broadly applicable in any research setting.
Keywords: insertion mutagenesis; protein fusions.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures






Similar articles
-
Libraries of green fluorescent protein fusions generated by transposition in vitro.Gene. 1998 Nov 19;222(2):213-22. doi: 10.1016/s0378-1119(98)00503-4. Gene. 1998. PMID: 9831655
-
New Mos1 mariner transposons suitable for the recovery of gene fusions in vivo and in vitro.Gene. 2001 Dec 12;280(1-2):97-105. doi: 10.1016/s0378-1119(01)00779-x. Gene. 2001. PMID: 11738822
-
Engineering green fluorescent protein as a dual functional tag.Biotechnol Bioeng. 2004 Jun 20;86(6):687-97. doi: 10.1002/bit.20077. Biotechnol Bioeng. 2004. PMID: 15137081
-
A new way to rapidly create functional, fluorescent fusion proteins: random insertion of GFP with an in vitro transposition reaction.BMC Neurosci. 2002 Jun 19;3:7. doi: 10.1186/1471-2202-3-7. BMC Neurosci. 2002. PMID: 12086589 Free PMC article.
-
Insertional gene fusion technology.FEBS Lett. 1999 Aug 20;457(1):1-4. doi: 10.1016/s0014-5793(99)00991-6. FEBS Lett. 1999. PMID: 10486551 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
