Influence of host phylogeny and water physicochemistry on microbial assemblages of the fish skin microbiome
- PMID: 38366921
- PMCID: PMC10903987
- DOI: 10.1093/femsec/fiae021
Influence of host phylogeny and water physicochemistry on microbial assemblages of the fish skin microbiome
Abstract
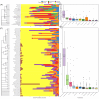
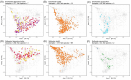
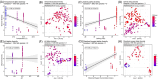
The skin of fish contains a diverse microbiota that has symbiotic functions with the host, facilitating pathogen exclusion, immune system priming, and nutrient degradation. The composition of fish skin microbiomes varies across species and in response to a variety of stressors, however, there has been no systematic analysis across these studies to evaluate how these factors shape fish skin microbiomes. Here, we examined 1922 fish skin microbiomes from 36 studies that included 98 species and nine rearing conditions to investigate associations between fish skin microbiome, fish species, and water physiochemical factors. Proteobacteria, particularly the class Gammaproteobacteria, were present in all marine and freshwater fish skin microbiomes. Acinetobacter, Aeromonas, Ralstonia, Sphingomonas and Flavobacterium were the most abundant genera within freshwater fish skin microbiomes, and Alteromonas, Photobacterium, Pseudoalteromonas, Psychrobacter and Vibrio were the most abundant in saltwater fish. Our results show that different culturing (rearing) environments have a small but significant effect on the skin bacterial community compositions. Water temperature, pH, dissolved oxygen concentration, and salinity significantly correlated with differences in beta-diversity but not necessarily alpha-diversity. To improve study comparability on fish skin microbiomes, we provide recommendations for approaches to the analyses of sequencing data and improve study reproducibility.
Keywords: 16S; V4; aquaculture; meta-analysis; microbiota; phylosymbiosis; physicochemical factors.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Akter T, Foysal MdJ, Alam M et al. Involvement of Enterococcus species in streptococcosis of Nile tilapia in Bangladesh. Aquaculture. 2021;531:735790. 10.1016/j.aquaculture.2020.735790. - DOI
-
- Arbizu PM. pairwiseAdonis. GitHub, 2018. https://github.com/pmartinezarbizu/pairwiseAdonis (8 February 2023, date last accessed).
-
- Austin B, Stuckey LF, Robertson PAW et al. A probiotic strain of Vibrio alginolyticus effective in reducing diseases caused by Aeromonas salmonicida, Vibrio anguillarum and Vibrio ordalii. J Fish Dis. 1995;18:93–6. 10.1111/j.1365-2761.1995.tb01271.x. - DOI

