The Greater Celandine: Identification and Characterization of an Antimicrobial Peptide from Chelidonium majus
- PMID: 38366995
- PMCID: PMC10959680
- DOI: 10.1021/acs.jnatprod.3c00939
The Greater Celandine: Identification and Characterization of an Antimicrobial Peptide from Chelidonium majus
Abstract
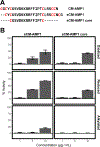
Chelidonium majus, known as Greater Celandine, is a latex-bearing plant that has been leveraged for its anticancer and antimicrobial properties. Herein, C. majus aerial tissue is mined for the presence of antimicrobial peptides. A highly abundant cysteine-rich peptide with a length of 25 amino acids, deemed CM-AMP1, is characterized through multiple mass spectrometric approaches. Electron-activated dissociation is leveraged to differentiate between isoleucine and leucine residues and complement conventional collision-induced dissociation to gain full sequence coverage of the full-length peptide. CM-AMP1 shares little sequence similarity with any proteins in publicly available databases, highlighting the novelty of its cysteine landscape and core motif. The presence of three disulfide bonds in the native peptide confers proteolytic stability, and antimicrobial activity is greatly decreased upon the alkylation of the cysteine residues. Synthetic variants of CM-AMP1 are used to confirm the activity of the full-length sequence and the core motif. To assess the biological impact, E. coli was grown in a sublethal concentration of CM-AMP1 and quantitative proteomics was used to identify proteins produced by the bacteria under stress, ultimately suggesting a membrane lytic antimicrobial mechanism of action. This study integrates multiple analytical methods for molecular and biological characterization of a unique antimicrobial peptide identified from C. majus.
Figures





References
-
- Zhang LJ; Gallo RL, Antimicrobial peptides. Curr Biol 2016, 26 (1), R14–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

