Identification of myoferlin as a mitochondria-associated membranes component required for calcium signaling in PDAC cell lines
- PMID: 38368370
- PMCID: PMC10874564
- DOI: 10.1186/s12964-024-01514-z
Identification of myoferlin as a mitochondria-associated membranes component required for calcium signaling in PDAC cell lines
Abstract
Background: Pancreatic ductal adenocarcinoma is an aggressive cancer type with one of the lowest survival rates due to late diagnosis and the absence of effective treatments. A better understanding of PDAC biology will help researchers to discover the Achilles' heel of cancer cells. In that regard, our research team investigated the function of an emerging oncoprotein known as myoferlin. Myoferlin is overexpressed in PDAC and its silencing/targeting has been shown to affect cancer cell proliferation, migration, mitochondrial dynamics and metabolism. Nevertheless, our comprehension of myoferlin functions in cells remains limited. In this study, we aimed to understand the molecular mechanism linking myoferlin silencing to mitochondrial dynamics.
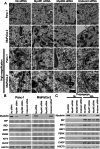
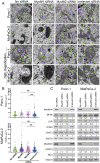
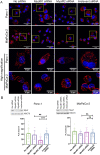
Methods: Experiments were performed on two pancreas cancer cell lines, Panc-1 and MiaPaCa-2. Myoferlin localization on mitochondria was evaluated by immunofluorescence, proximity ligation assay, and cell fractionation. The presence of myoferlin in mitochondria-associated membranes was assessed by cell fractionation and its function in mitochondrial calcium transfer was evaluated using calcium flow experiments, proximity ligation assays, co-immunoprecipitation, and timelapse fluorescence microscopy in living cells.
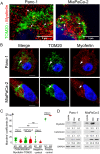
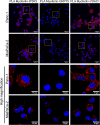
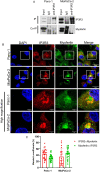
Results: Myoferlin localization on mitochondria was investigated. Our results suggest that myoferlin is unlikely to be located on mitochondria. Instead, we identified myoferlin as a new component of mitochondria-associated membranes. Its silencing significantly reduces the mitochondrial calcium level upon stimulation, probably through myoferlin interaction with the inositol 1,4,5-triphosphate receptors 3.
Conclusions: For the first time, myoferlin was specifically demonstrated to be located in mitochondria-associated membranes where it participates to calcium flow. We hypothesized that this function explains our previous results on mitochondrial dynamics. This study improves our comprehension of myoferlin localization and function in cancer biology.
Keywords: Calcium signaling; ER-mitochondria contact sites; IP3R3; Mitochondria; Mitochondria-associated membranes; Myoferlin; Pancreatic cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

