Concentration strategies for spiked and naturally present biomarkers in non-invasively collected first-void urine
- PMID: 38368382
- PMCID: PMC10873940
- DOI: 10.1186/s40001-024-01719-5
Concentration strategies for spiked and naturally present biomarkers in non-invasively collected first-void urine
Abstract
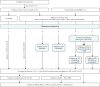
Background: First-void urine (FVU) provides a non-invasive method for collecting a wide range of biomarkers found in genital tract secretions. To optimize biomarker collection in FVU, this study investigated the impact of naturally present and supplemented precipitating agents: uromodulin (UMOD) and polyethylene glycol (PEG), on the concentration of human papillomavirus (HPV) pseudovirions (PsV), cell-free DNA (cfDNA), and cellular genomic DNA (gDNA) through centrifugation.
Methods: FVU samples from ten healthy female volunteers, along with a control sample, were spiked with seal herpesvirus 1 (PhHV-1) DNA, HPV16 plasmid DNA, and HPV16 PsV with an enhanced green fluorescent protein (EGFP) reporter. The samples were subjected to various concentration protocols involving PEG precipitation, low-speed centrifugation (5 min at 1000×g), and medium-speed centrifugation (1 h at 3000×g). Subsequently, quantitative PCR (qPCR) was used to assess cellular and cell-free glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA, cell-free PhHV-1 and HPV16 DNA, and PsV (EGFP) DNA. In addition, UMOD levels were measured.
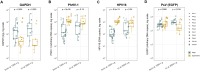
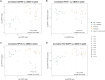
Results: The findings revealed that PEG significantly increased the concentration of cfDNA and gDNA in the pellet after centrifugation, with the most pronounced effect observed for cfDNA. Moreover, low-speed centrifugation without PEG effectively depleted cellular gDNA while preserving cfDNA in the supernatants. Pseudovirions were consistently pelleted, even with low-speed centrifugation, and a positive but not significant effect of PEG on PsV (EGFP) DNA yield in the pellet was observed. Additionally, a significant correlation was observed between UMOD and GAPDH, HPV16, and PsV (EGFP) DNA quantities in the pellet. Furthermore, large variations among the FVU samples were observed.
Conclusions: With this study, we provide novel insights into how various biomarker precipitation protocols, including both the properties of FVU and the use of PEG as a precipitating agent, influence the concentration of cfDNA, cellular gDNA, and pseudovirions.
Keywords: Biomarkers; Concentration; FVU; First-void urine; Human papillomavirus.
© 2024. The Author(s).
Conflict of interest statement
A.V. is a co-founder and former board member of Novosanis (Subsidiary of OraSure Technologies Inc, Wijnegem, Belgium), a spin-off company of the University of Antwerp, and was a minority shareholder until January 2019. The University of Antwerp received grants from Merck, GSK, Hologic, Abbott, Roche, and Cepheid to support the HPV Prevention and Control Board. The University of Antwerp received a project grant and honoraria fee for lectures, presentations, and speaker bureaus from Merck. Other authors declare that they have no conflict of interest.
Figures

Similar articles
-
HPV-specific antibodies in female genital tract secretions captured via first-void urine retain their neutralizing capacity.Hum Vaccin Immunother. 2024 Dec 31;20(1):2330168. doi: 10.1080/21645515.2024.2330168. Epub 2024 Apr 3. Hum Vaccin Immunother. 2024. PMID: 38567541 Free PMC article.
-
Stability, enrichment, and quantification of total and HPV16-specific IgG present in first-void urine.Sci Rep. 2024 Jun 23;14(1):14441. doi: 10.1038/s41598-024-65257-0. Sci Rep. 2024. PMID: 38910149 Free PMC article.
-
Impact of Collection Volume and DNA Extraction Method on the Detection of Biomarkers and HPV DNA in First-Void Urine.Molecules. 2021 Apr 1;26(7):1989. doi: 10.3390/molecules26071989. Molecules. 2021. PMID: 33915837 Free PMC article.
-
First-void urine: A potential biomarker source for triage of high-risk human papillomavirus infected women.Eur J Obstet Gynecol Reprod Biol. 2017 Sep;216:1-11. doi: 10.1016/j.ejogrb.2017.06.036. Epub 2017 Jun 27. Eur J Obstet Gynecol Reprod Biol. 2017. PMID: 28689156 Review.
-
cfDNA detection for HPV+ squamous cell carcinomas.Oral Oncol. 2021 Apr;115:104958. doi: 10.1016/j.oraloncology.2020.104958. Epub 2021 Feb 11. Oral Oncol. 2021. PMID: 33582486 Free PMC article. Review.
Cited by
-
HPV-specific antibodies in female genital tract secretions captured via first-void urine retain their neutralizing capacity.Hum Vaccin Immunother. 2024 Dec 31;20(1):2330168. doi: 10.1080/21645515.2024.2330168. Epub 2024 Apr 3. Hum Vaccin Immunother. 2024. PMID: 38567541 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

