COVID-19 immune signatures in Uganda persist in HIV co-infection and diverge by pandemic phase
- PMID: 38368384
- PMCID: PMC10874401
- DOI: 10.1038/s41467-024-45204-3
COVID-19 immune signatures in Uganda persist in HIV co-infection and diverge by pandemic phase
Abstract
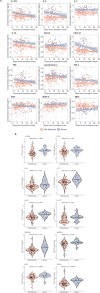
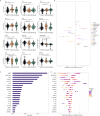
Little is known about the pathobiology of SARS-CoV-2 infection in sub-Saharan Africa, where severe COVID-19 fatality rates are among the highest in the world and the immunological landscape is unique. In a prospective cohort study of 306 adults encompassing the entire clinical spectrum of SARS-CoV-2 infection in Uganda, we profile the peripheral blood proteome and transcriptome to characterize the immunopathology of COVID-19 across multiple phases of the pandemic. Beyond the prognostic importance of myeloid cell-driven immune activation and lymphopenia, we show that multifaceted impairment of host protein synthesis and redox imbalance define core biological signatures of severe COVID-19, with central roles for IL-7, IL-15, and lymphotoxin-α in COVID-19 respiratory failure. While prognostic signatures are generally consistent in SARS-CoV-2/HIV-coinfection, type I interferon responses uniquely scale with COVID-19 severity in persons living with HIV. Throughout the pandemic, COVID-19 severity peaked during phases dominated by A.23/A.23.1 and Delta B.1.617.2/AY variants. Independent of clinical severity, Delta phase COVID-19 is distinguished by exaggerated pro-inflammatory myeloid cell and inflammasome activation, NK and CD8+ T cell depletion, and impaired host protein synthesis. Combining these analyses with a contemporary Ugandan cohort of adults hospitalized with influenza and other severe acute respiratory infections, we show that activation of epidermal and platelet-derived growth factor pathways are distinct features of COVID-19, deepening translational understanding of mechanisms potentially underlying SARS-CoV-2-associated pulmonary fibrosis. Collectively, our findings provide biological rationale for use of broad and targeted immunotherapies for severe COVID-19 in sub-Saharan Africa, illustrate the relevance of local viral and host factors to SARS-CoV-2 immunopathology, and highlight underemphasized yet therapeutically exploitable immune pathways driving COVID-19 severity.
© 2024. The Author(s).
Conflict of interest statement
M.J.C. and M.R.O’D. were investigators for clinical trials evaluating the efficacy and safety of remdesivir, convalescent plasma, and anti-SARS-CoV-2 hyperimmune globulin in hospitalized patients with COVID-19, sponsored by Gilead Sciences, Amazon, and the National Institutes of Health, respectively. Compensation for this work was paid to Columbia University. The remaining authors declare no competing interests.
Figures





References
-
- US Centers for Disease Control and Prevention. COVID-19 Data Review: update on COVID-19–related mortality. https://www.cdc.gov/coronavirus/2019-ncov/science/data-review/index.html. (2023).
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

