Traditional potato tillage systems in the Peruvian Andes impact bacterial diversity, evenness, community composition, and functions in soil microbiomes
- PMID: 38368478
- PMCID: PMC10874408
- DOI: 10.1038/s41598-024-54652-2
Traditional potato tillage systems in the Peruvian Andes impact bacterial diversity, evenness, community composition, and functions in soil microbiomes
Abstract
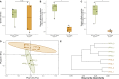
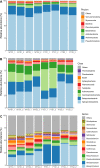
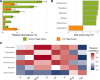
The soil microbiome, a crucial component of agricultural ecosystems, plays a pivotal role in crop production and ecosystem functioning. However, its response to traditional tillage systems in potato cultivation in the Peruvian highlands is still far from understood. Here, ecological and functional aspects of the bacterial community were analyzed based on soil samples from two traditional tillage systems: 'chiwa' (minimal tillage) and 'barbecho' (full tillage), in the Huanuco region of the Peruvian central Andes. Similar soil bacterial community composition was shown for minimal tillage system, but it was heterogeneous for full tillage system. This soil bacterial community composition under full tillage system may be attributed to stochastic, and a more dynamic environment within this tillage system. 'Chiwa' and 'barbecho' soils harbored distinct bacterial genera into their communities, indicating their potential as bioindicators of traditional tillage effects. Functional analysis revealed common metabolic pathways in both tillage systems, with differences in anaerobic pathways in 'chiwa' and more diverse pathways in 'barbecho'. These findings open the possibilities to explore microbial bioindicators for minimal and full tillage systems, which are in relationship with healthy soil, and they can be used to propose adequate tillage systems for the sowing of potatoes in Peru.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Structure, composition and metagenomic profile of soil microbiomes associated to agricultural land use and tillage systems in Argentine Pampas.PLoS One. 2014 Jun 12;9(6):e99949. doi: 10.1371/journal.pone.0099949. eCollection 2014. PLoS One. 2014. PMID: 24923965 Free PMC article.
-
Temporal Dynamics of Bacterial Communities along a Gradient of Disturbance in a U.S. Southern Plains Agroecosystem.mBio. 2022 Jun 28;13(3):e0382921. doi: 10.1128/mbio.03829-21. Epub 2022 Apr 14. mBio. 2022. PMID: 35420482 Free PMC article.
-
Effects of tillage practices on soil microbiome and agricultural parameters.Sci Total Environ. 2020 Feb 25;705:135791. doi: 10.1016/j.scitotenv.2019.135791. Epub 2019 Nov 27. Sci Total Environ. 2020. PMID: 31810706
-
Potato tillage method is associated with soil microbial communities, soil chemical properties, and potato yield.J Microbiol. 2022 Feb;60(2):156-166. doi: 10.1007/s12275-022-1060-0. Epub 2022 Jan 7. J Microbiol. 2022. PMID: 34994959
-
No tillage and residue mulching method on bacterial community diversity regulation in a black soil region of Northeastern China.PLoS One. 2021 Sep 10;16(9):e0256970. doi: 10.1371/journal.pone.0256970. eCollection 2021. PLoS One. 2021. PMID: 34506513 Free PMC article.
Cited by
-
Sandy loam soil maintains better physicochemical parameters and more abundant beneficial microbiomes than clay soil in Stevia rebaudiana cultivation.PeerJ. 2024 Sep 19;12:e18010. doi: 10.7717/peerj.18010. eCollection 2024. PeerJ. 2024. PMID: 39308829 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

