Improving cryo-EM grids for amyloid fibrils using interface-active solutions and spectator proteins
- PMID: 38368506
- PMCID: PMC10995402
- DOI: 10.1016/j.bpj.2024.02.009
Improving cryo-EM grids for amyloid fibrils using interface-active solutions and spectator proteins
Abstract
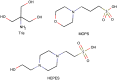
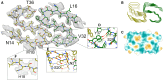
Preparation of cryoelectron microscopy (cryo-EM) grids for imaging of amyloid fibrils is notoriously challenging. The human islet amyloid polypeptide (hIAPP) serves as a notable example, as the majority of reported structures have relied on the use of nonphysiological pH buffers, N-terminal tags, and seeding. This highlights the need for more efficient, reproducible methodologies that can elucidate amyloid fibril structures formed under diverse conditions. In this work, we demonstrate that the distribution of fibrils on cryo-EM grids is predominantly determined by the solution composition, which is critical for the stability of thin vitreous ice films. We discover that, among physiological pH buffers, HEPES uniquely enhances the distribution of fibrils on cryo-EM grids and improves the stability of ice layers. This improvement is attributed to direct interactions between HEPES molecules and hIAPP, effectively minimizing the tendency of hIAPP to form dense clusters in solutions and preventing ice nucleation. Furthermore, we provide additional support for the idea that denatured protein monolayers forming at the interface are also capable of eliciting a surfactant-like effect, leading to improved particle coverage. This phenomenon is illustrated by the addition of nonamyloidogenic rat IAPP (rIAPP) to a solution of preaggregated hIAPP just before the freezing process. The resultant grids, supplemented with this "spectator protein", exhibit notably enhanced coverage and improved ice quality. Unlike conventional surfactants, rIAPP is additionally capable of disentangling the dense clusters formed by hIAPP. By applying the proposed strategies, we have resolved the structure of the dominant hIAPP polymorph, formed in vitro at pH 7.4, to a final resolution of 4 Å. The advances in grid preparation presented in this work hold significant promise for enabling structural determination of amyloid proteins which are particularly resistant to conventional grid preparation techniques.
Copyright © 2024 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Similar articles
-
Cryo-Electron Microscopy Provides Mechanistic Insights into Solution-Dependent Polymorphism and Cross-Aggregation Phenomena of the Human and Rat Islet Amyloid Polypeptides.Biochemistry. 2025 Jun 17;64(12):2583-2595. doi: 10.1021/acs.biochem.5c00042. Epub 2025 May 26. Biochemistry. 2025. PMID: 40417836 Free PMC article.
-
Structural and energetic insight into the cross-seeding amyloid assemblies of human IAPP and rat IAPP.J Phys Chem B. 2014 Jun 26;118(25):7026-36. doi: 10.1021/jp5022246. Epub 2014 Jun 12. J Phys Chem B. 2014. PMID: 24892388
-
Cryo-EM structures of hIAPP fibrils seeded by patient-extracted fibrils reveal new polymorphs and conserved fibril cores.Nat Struct Mol Biol. 2021 Sep;28(9):724-730. doi: 10.1038/s41594-021-00646-x. Epub 2021 Sep 9. Nat Struct Mol Biol. 2021. PMID: 34518699 Free PMC article.
-
Characterisation of the Structure and Oligomerisation of Islet Amyloid Polypeptides (IAPP): A Review of Molecular Dynamics Simulation Studies.Molecules. 2018 Aug 25;23(9):2142. doi: 10.3390/molecules23092142. Molecules. 2018. PMID: 30149632 Free PMC article. Review.
-
Islet amyloid polypeptide: aggregation and fibrillogenesis in vitro and its inhibition.Subcell Biochem. 2012;65:185-209. doi: 10.1007/978-94-007-5416-4_8. Subcell Biochem. 2012. PMID: 23225004 Review.
Cited by
-
The Structure of Human IAPP Fibrils Reflects Membrane and pH Conditions.J Am Chem Soc. 2025 Aug 13;147(32):28943-28954. doi: 10.1021/jacs.5c06971. Epub 2025 Aug 1. J Am Chem Soc. 2025. PMID: 40748680 Free PMC article.
-
Cryo-Electron Microscopy Provides Mechanistic Insights into Solution-Dependent Polymorphism and Cross-Aggregation Phenomena of the Human and Rat Islet Amyloid Polypeptides.Biochemistry. 2025 Jun 17;64(12):2583-2595. doi: 10.1021/acs.biochem.5c00042. Epub 2025 May 26. Biochemistry. 2025. PMID: 40417836 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

