Unveiling the microbial communities and metabolic pathways of Keem, a traditional starter culture, through whole-genome sequencing
- PMID: 38369518
- PMCID: PMC10874962
- DOI: 10.1038/s41598-024-53350-3
Unveiling the microbial communities and metabolic pathways of Keem, a traditional starter culture, through whole-genome sequencing
Abstract
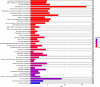
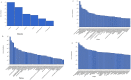
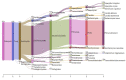
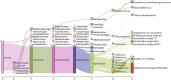
Traditional alcoholic beverages have played a significant role in the cultural, social, and culinary fabric of societies worldwide for centuries. Studying the microbial community structure and their metabolic potential in such beverages is necessary to define product quality, safety, and consistency, as well as to explore associated biotechnological applications. In the present investigation, Illumina-based (MiSeq system) whole-genome shotgun sequencing was utilized to characterize the microbial diversity and conduct predictive gene function analysis of keem, a starter culture employed by the Jaunsari tribal community in India for producing various traditional alcoholic beverages. A total of 8,665,213 sequences, with an average base length of 151 bps, were analyzed using MG-RAST. The analysis revealed the dominance of bacteria (95.81%), followed by eukaryotes (4.11%), archaea (0.05%), and viruses (0.03%). At the phylum level, Actinobacteria (81.18%) was the most abundant, followed by Firmicutes (10.56%), Proteobacteria (4.00%), and Ascomycota (3.02%). The most predominant genera were Saccharopolyspora (36.31%), followed by Brevibacterium (15.49%), Streptomyces (9.52%), Staphylococcus (8.75%), Bacillus (4.59%), and Brachybacterium (3.42%). At the species level, the bacterial, fungal, and viral populations of the keem sample could be categorized into 3347, 57, and 106 species, respectively. Various functional attributes to the sequenced data were assigned using Cluster of Orthologous Groups (COG), Non-supervised Orthologous Groups (NOG), subsystem, and KEGG Orthology (KO) annotations. The most prevalent metabolic pathways included carbohydrate, lipid, and amino acid metabolism, as well as the biosynthesis of glycans, secondary metabolites, and xenobiotic biodegradation. Given the rich microbial diversity and its associated metabolic potential, investigating the transition of keem from a traditional starter culture to an industrial one presents a compelling avenue for future research.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Microbial Diversity and Functional Potential of Keem: A Traditional Starter Culture for Alcoholic Beverage-Application of Next-Generation Amplicon and Shotgun Metagenome Sequences.Mol Biotechnol. 2023 Aug 11. doi: 10.1007/s12033-023-00839-3. Online ahead of print. Mol Biotechnol. 2023. PMID: 37566190
-
Epilithic Biofilms in Lake Baikal: Screening and Diversity of PKS and NRPS Genes in the Genomes of Heterotrophic Bacteria.Pol J Microbiol. 2018;67(4):501-516. doi: 10.21307/pjm-2018-060. Pol J Microbiol. 2018. PMID: 30550237 Free PMC article.
-
Deciphering the microbiota data from termite mound soil in South Africa using shotgun metagenomics.Data Brief. 2019 Nov 13;28:104802. doi: 10.1016/j.dib.2019.104802. eCollection 2020 Feb. Data Brief. 2019. PMID: 31832528 Free PMC article.
-
Structural and functional characteristics of soil microbial community in a Pinus massoniana forest at different elevations.PeerJ. 2022 Jul 15;10:e13504. doi: 10.7717/peerj.13504. eCollection 2022. PeerJ. 2022. PMID: 35860041 Free PMC article.
-
Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review.Microorganisms. 2021 Jul 9;9(7):1473. doi: 10.3390/microorganisms9071473. Microorganisms. 2021. PMID: 34361908 Free PMC article. Review.
Cited by
-
Highlighting Lactic Acid Bacteria in Beverages: Diversity, Fermentation, Challenges, and Future Perspectives.Foods. 2025 Jun 10;14(12):2043. doi: 10.3390/foods14122043. Foods. 2025. PMID: 40565651 Free PMC article. Review.
References
-
- Thakur N, Bhalla TC. Characterization of some traditional fermented foods and beverages of Himachal Pradesh. Indian J. Tradit. Knowl. 2004;3(3):325–335.
-
- Steinkraus KH. Industrialization of Indigenous Fermented Foods, Basel. Marcel Dekker Inc.; 1989.
-
- Tamang JP. Diversity of fermented beverages and alcoholic drinks. In: Tamang JP, Kailasapathy K, editors. Fermented Foods and Beverages of the World. CRC Press; 2010. pp. 85–126.
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

