Impact of the diet in the gut microbiota after an inter-species microbial transplantation in fish
- PMID: 38369563
- PMCID: PMC10874947
- DOI: 10.1038/s41598-024-54519-6
Impact of the diet in the gut microbiota after an inter-species microbial transplantation in fish
Abstract
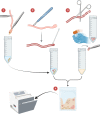
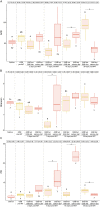
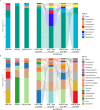
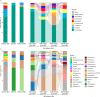
Inter-species microbial transplantations offer the possibility of transferring species-specific microbes and their associated functionality. As a conceptual approach, an intestinal microbiota transplant (IMT) between two marine carnivorous fish species that thrive in different environmental conditions was conducted: from donor Atlantic salmon (Salmo salar) to recipient gilthead seabream (Sparus aurata), after obliterating its basal microbiota with an antibiotic treatment. To confirm that the gut microbiota was able to recover after antibiotics without the influence of the diet, a group of gilthead seabream not submitted to the IMT was kept fasted as an internal control. To assess the effect of the diet after the IMT, two groups of gilthead seabream were respectively fed with their typical diet and with Atlantic salmon diet. At 36 days post-IMT, the gut of the individuals fed with their typical diet was dominated by the feed-associated bacteria, while those fed with the salmon diet had developed a unique microbiota from the convergence of the diet, donor, and recipient microbiota. These results suggested that an intestinal microbiota transplantation may be effective if the basal microbiota from the gut is first cleared and a targeted dietary modification is provided to maintain and enrich the novel bacteria species over time.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

