Modeling the SDF-1/CXCR4 protein using advanced artificial intelligence and antagonist screening for Japanese anchovy
- PMID: 38370015
- PMCID: PMC10869568
- DOI: 10.3389/fphys.2024.1349119
Modeling the SDF-1/CXCR4 protein using advanced artificial intelligence and antagonist screening for Japanese anchovy
Abstract
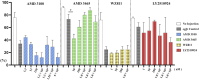
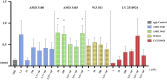
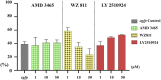
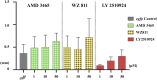
SDF-1/CXCR4 chemokine signaling are indispensable for cell migration, especially the Primordial Germ Cell (PGC) migration towards the gonadal ridge during early development. We earlier found that this signaling is largely conserved in the Japanese anchovy (Engraulis japonicus, EJ), and a mere treatment of CXCR4 antagonist, AMD3100, leads to germ cell depletion and thereafter gonad sterilization. However, the effect of AMD3100 was limited. So, in this research, we scouted for CXCR4 antagonist with higher potency by employing advanced artificial intelligence deep learning-based computer simulations. Three potential candidates, AMD3465, WZ811, and LY2510924, were selected and in vivo validation was conducted using Japanese anchovy embryos. We found that seven transmembrane motif of EJ CXCR4a and EJ CXCR4b were extremely similar with human homolog while the CXCR4 chemokine receptor N terminal (PF12109, essential for SDF-1 binding) was missing in EJ CXCR4b. 3D protein analysis and cavity search predicted the cavity in EJ CXCR4a to be five times larger (6,307 ų) than that in EJ CXCR4b (1,241 ų). Docking analysis demonstrated lower binding energy of AMD3100 and AMD3465 to EJ CXCR4a (Vina score -9.6) and EJ CXCR4b (Vina score -8.8), respectively. Furthermore, we observed significant PGC mismigration in microinjected AMD3465 treated groups at 10, 100 and 1 × 105 nM concentration in 48 h post fertilized embryos. The other three antagonists showed various degrees of PGC dispersion, but no significant effect compared to their solvent control at tested concentrations was observed. Cumulatively, our results suggests that AMD3645 might be a better candidate for abnormal PGC migration in Japanese anchovy and warrants further investigation.
Keywords: Alpha Fold; CB Dock; Colab Fold; Japanese anchovy; PGC; SDF-1/CXCR4.
Copyright © 2024 Yahiro, Barnuevo, Sato, Mohapatra, Toyoda, Itoh, Ohno, Matsuyama, Chakraborty and Ohta.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
SDF-1/CXCR4 signal is involved in the induction of Primordial Germ Cell migration in a model marine fish, Japanese anchovy (Engraulis japonicus).Gen Comp Endocrinol. 2024 May 15;351:114476. doi: 10.1016/j.ygcen.2024.114476. Epub 2024 Feb 24. Gen Comp Endocrinol. 2024. PMID: 38408712
-
Analysis of SDF-1/CXCR4 signaling in primordial germ cell migration and survival or differentiation in Xenopus laevis.Mech Dev. 2010 Jan-Feb;127(1-2):146-58. doi: 10.1016/j.mod.2009.09.005. Epub 2009 Sep 19. Mech Dev. 2010. PMID: 19770040
-
Expression and potential role of chemokine receptor CXCR4 in human bladder carcinoma cell lines with different metastatic ability.Mol Med Rep. 2011 May-Jun;4(3):525-8. doi: 10.3892/mmr.2011.440. Epub 2011 Feb 18. Mol Med Rep. 2011. PMID: 21468602
-
MSX-122: Is an effective small molecule CXCR4 antagonist in cancer therapy?Int Immunopharmacol. 2022 Jul;108:108863. doi: 10.1016/j.intimp.2022.108863. Epub 2022 May 25. Int Immunopharmacol. 2022. PMID: 35623288 Review.
-
[Some research progress of CXCR4 antagonist AMD3100].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2011 Jun;19(3):831-4. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2011. PMID: 21729582 Review. Chinese.
References
-
- Ali P. K. M. M., Rao G. P. S. (1989). Growth improvement in carp, Cyprinus carpio (Linnaeus), sterilized with 17α-methyltestosterone. Aquaculture 76, 157–167. 10.1016/0044-8486(89)90260-3 - DOI
-
- Crump M. P., Gong J.-H., Loetscher P., Rajarathnam K., Amara A., Arenzana-Seisdedos F., et al. (1997). Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 16, 6996–7007. 10.1093/emboj/16.23.6996 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

