Exploring candidate genes for heat tolerance in ovine through liver gene expression
- PMID: 38370230
- PMCID: PMC10869868
- DOI: 10.1016/j.heliyon.2024.e25692
Exploring candidate genes for heat tolerance in ovine through liver gene expression
Abstract
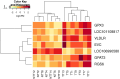
Thermotolerance has become an essential factor in the prevention of the adverse effects of heat stress, but it varies among animals. Identifying genes related to heat adaptability traits is important for improving thermotolerance and for selecting more productive animals in hot environments. The primary objective of this research was to find candidate genes in the liver that play a crucial role in the heat stress response of Santa Ines sheep, which exhibit varying levels of heat tolerance. To achieve this goal, 80 sheep were selected based on their thermotolerance and placed in a climate chamber for 10 days, during which the average temperature was maintained at 36 °C from 10 a.m. to 4 p.m. and 28 °C from 4 p.m. to 10 a.m. A subset of 14 extreme animals, with seven thermotolerant and seven non-thermotolerant animals based on heat loss (rectal temperature), were selected for liver sampling. RNA sequencing and differential gene expression analysis were performed. Thermotolerant sheep showed higher expression of genes GPx3, RGS6, GPAT3, VLDLR, LOC101108817, and EVC. These genes were mainly related to the Hedgehog signaling pathway, glutathione metabolism, glycerolipid metabolism, and thyroid hormone synthesis. These enhanced pathways in thermotolerant animals could potentially mitigate the negative effects of heat stress, conferring greater heat resistance.
Keywords: Gene; Sheep; Thermotolerance; Transcriptomic.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Proteomic identification of potential biomarkers for heat tolerance in Caracu beef cattle using high and low thermotolerant groups.BMC Genomics. 2024 Nov 13;25(1):1079. doi: 10.1186/s12864-024-11021-7. BMC Genomics. 2024. PMID: 39538142 Free PMC article.
-
Skin transcriptomic analysis reveals candidate genes and pathways associated with thermotolerance in hair sheep.Int J Biometeorol. 2024 Mar;68(3):435-444. doi: 10.1007/s00484-023-02602-4. Epub 2023 Dec 26. Int J Biometeorol. 2024. PMID: 38147121
-
The ability to induce heat shock transcription factor-regulated genes in response to lethal heat stress is associated with thermotolerance in tomato cultivars.Front Plant Sci. 2023 Oct 5;14:1269964. doi: 10.3389/fpls.2023.1269964. eCollection 2023. Front Plant Sci. 2023. PMID: 37868310 Free PMC article.
-
Impact of heat stress on dairy cattle and selection strategies for thermotolerance: a review.Front Vet Sci. 2023 Jun 20;10:1198697. doi: 10.3389/fvets.2023.1198697. eCollection 2023. Front Vet Sci. 2023. PMID: 37408833 Free PMC article. Review.
-
Effects of Heat stress and molecular mitigation approaches in orphan legume, Chickpea.Mol Biol Rep. 2020 Jun;47(6):4659-4670. doi: 10.1007/s11033-020-05358-x. Epub 2020 Mar 4. Mol Biol Rep. 2020. PMID: 32133603 Review.
Cited by
-
Potential Genetic Markers Associated with Environmental Adaptability in Herbivorous Livestock.Animals (Basel). 2025 Mar 5;15(5):748. doi: 10.3390/ani15050748. Animals (Basel). 2025. PMID: 40076029 Free PMC article. Review.
-
Proteomic identification of potential biomarkers for heat tolerance in Caracu beef cattle using high and low thermotolerant groups.BMC Genomics. 2024 Nov 13;25(1):1079. doi: 10.1186/s12864-024-11021-7. BMC Genomics. 2024. PMID: 39538142 Free PMC article.
References
-
- Molaei Moghbeli S., Barazandeh A., Vatankhah M., Mohammadabadi M. Genetics and non-genetics parameters of body weight for post-weaning traits in Raini Cashmere goats. Trop. Anim. Health Prod. 2013;45:1519–1524. - PubMed
-
- Oliveira F.G., Sousa W.H., Cartaxo F.Q., Batista A.S.M., Ramos J.P.F., Cavalcante I.T.R. Quality of meat from Santa Ines sheep with different biotypes and slaughtering weights. Rev. Bras. Saude Prod. Anim. 2020;21:1–13. doi: 10.1590/s1519-994020210732020. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

