This is a preprint.
A meta-analysis of the gut microbiome in inflammatory bowel disease patients identifies disease-associated small molecules
- PMID: 38370680
- PMCID: PMC10871352
- DOI: 10.1101/2024.02.07.579278
A meta-analysis of the gut microbiome in inflammatory bowel disease patients identifies disease-associated small molecules
Update in
-
A meta-analysis of the gut microbiome in inflammatory bowel disease patients identifies disease-associated small molecules.Cell Host Microbe. 2025 Feb 12;33(2):218-234.e12. doi: 10.1016/j.chom.2025.01.002. Cell Host Microbe. 2025. PMID: 39947133
Abstract
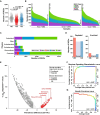
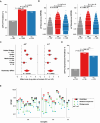
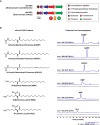
Changes in the gut microbiome have been associated with several human diseases, but the molecular and functional details underlying these associations remain largely unknown. Here, we performed a multi-cohort analysis of small molecule biosynthetic gene clusters (BGCs) in 5,306 metagenomic samples of the gut microbiome from 2,033 Inflammatory Bowel Disease (IBD) patients and 833 matched healthy subjects and identified a group of Clostridia-derived BGCs that are significantly associated with IBD. Using synthetic biology, we discovered and solved the structures of six fatty acid amides as the products of the IBD-enriched BGCs. Using two mouse models of colitis, we show that the discovered small molecules disrupt gut permeability and exacerbate inflammation in chemically and genetically susceptible mice. These findings suggest that microbiome-derived small molecules may play a role in the etiology of IBD and represent a generalizable approach for discovering molecular mediators of microbiome-host interactions in the context of microbiome-associated diseases.
Conflict of interest statement
Declaration of Interests M.S.D. is a Scientific Co-Founder and CSO at Pragma Biosciences.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
