This is a preprint.
A pharmacological toolkit for human microglia identifies Topoisomerase I inhibitors as immunomodulators for Alzheimer's disease
- PMID: 38370689
- PMCID: PMC10871172
- DOI: 10.1101/2024.02.06.579103
A pharmacological toolkit for human microglia identifies Topoisomerase I inhibitors as immunomodulators for Alzheimer's disease
Abstract
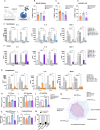
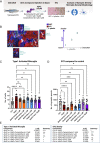
While efforts to identify microglial subtypes have recently accelerated, the relation of transcriptomically defined states to function has been largely limited to in silico annotations. Here, we characterize a set of pharmacological compounds that have been proposed to polarize human microglia towards two distinct states - one enriched for AD and MS genes and another characterized by increased expression of antigen presentation genes. Using different model systems including HMC3 cells, iPSC-derived microglia and cerebral organoids, we characterize the effect of these compounds in mimicking human microglial subtypes in vitro. We show that the Topoisomerase I inhibitor Camptothecin induces a CD74high/MHChigh microglial subtype which is specialized in amyloid beta phagocytosis. Camptothecin suppressed amyloid toxicity and restored microglia back to their homeostatic state in a zebrafish amyloid model. Our work provides avenues to recapitulate human microglial subtypes in vitro, enabling functional characterization and providing a foundation for modulating human microglia in vivo.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
