This is a preprint.
Protein-protein interactions with G3BPs drive stress granule condensation and gene expression changes under cellular stress
- PMID: 38370785
- PMCID: PMC10871250
- DOI: 10.1101/2024.02.06.579149
Protein-protein interactions with G3BPs drive stress granule condensation and gene expression changes under cellular stress
Update in
-
G3BP isoforms differentially affect stress granule assembly and gene expression during cellular stress.Mol Biol Cell. 2024 Nov 1;35(11):ar140. doi: 10.1091/mbc.E24-02-0062. Epub 2024 Oct 2. Mol Biol Cell. 2024. PMID: 39356796 Free PMC article.
Abstract
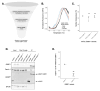
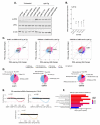
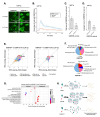
Stress granules (SGs) are macromolecular assemblies that form under cellular stress. Formation of these condensates is driven by the condensation of RNA and RNA-binding proteins such as G3BPs. G3BPs condense into SGs following stress-induced translational arrest. Three G3BP paralogs (G3BP1, G3BP2A, and G3BP2B) have been identified in vertebrates. However, the contribution of different G3BP paralogs to stress granule formation and stress-induced gene expression changes is incompletely understood. Here, we identified key residues for G3BP condensation such as V11. This conserved amino acid is required for formation of the G3BP-Caprin-1 complex, hence promoting SG assembly. Total RNA sequencing and ribosome profiling revealed that disruption of G3BP condensation corresponds to changes in mRNA levels and ribosome engagement during the integrated stress response (ISR). Moreover, we found that G3BP2B preferentially condenses and promotes changes in mRNA expression under endoplasmic reticulum (ER) stress. Together, this work suggests that stress granule assembly promotes changes in gene expression under cellular stress, which is differentially regulated by G3BP paralogs.
Keywords: G3BPs; Stress granules; integrated stress response; ribosome profiling.
Figures



Similar articles
-
G3BP isoforms differentially affect stress granule assembly and gene expression during cellular stress.Mol Biol Cell. 2024 Nov 1;35(11):ar140. doi: 10.1091/mbc.E24-02-0062. Epub 2024 Oct 2. Mol Biol Cell. 2024. PMID: 39356796 Free PMC article.
-
Pseudorabies virus IE180 protein hijacks G3BPs into the nucleus to inhibit stress granule formation.J Virol. 2025 Apr 15;99(4):e0208824. doi: 10.1128/jvi.02088-24. Epub 2025 Mar 27. J Virol. 2025. PMID: 40145738 Free PMC article.
-
G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits.J Cell Biol. 2016 Mar 28;212(7):845-60. doi: 10.1083/jcb.201508028. J Cell Biol. 2016. PMID: 27022092 Free PMC article.
-
Rasputin a decade on and more promiscuous than ever? A review of G3BPs.Biochim Biophys Acta Mol Cell Res. 2019 Mar;1866(3):360-370. doi: 10.1016/j.bbamcr.2018.09.001. Epub 2018 Sep 5. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30595162 Free PMC article. Review.
-
Role(s) of G3BPs in Human Pathogenesis.J Pharmacol Exp Ther. 2023 Oct;387(1):100-110. doi: 10.1124/jpet.122.001538. Epub 2023 Jul 19. J Pharmacol Exp Ther. 2023. PMID: 37468286 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
