This is a preprint.
A Simple and Scalable Zebrafish Model of Sonic Hedgehog Medulloblastoma
- PMID: 38370799
- PMCID: PMC10871209
- DOI: 10.1101/2024.02.03.577834
A Simple and Scalable Zebrafish Model of Sonic Hedgehog Medulloblastoma
Update in
-
A simple and scalable zebrafish model of Sonic hedgehog medulloblastoma.Cell Rep. 2024 Aug 27;43(8):114559. doi: 10.1016/j.celrep.2024.114559. Epub 2024 Jul 29. Cell Rep. 2024. PMID: 39078737 Free PMC article.
Abstract
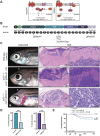
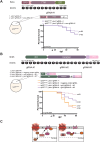
Medulloblastoma (MB) is the most common malignant brain tumor in children and is stratified into three major subgroups. The Sonic hedgehog (SHH) subgroup represents ~30% of all MB cases and has significant survival disparity depending upon TP53 status. Here, we describe the first zebrafish model of SHH MB using CRISPR to mutate ptch1, the primary genetic driver in human SHH MB. These tumors rapidly arise adjacent to the valvula cerebelli and resemble human SHH MB by histology and comparative genomics. In addition, ptch1-deficient MB tumors with loss of tp53 have aggressive tumor histology and significantly worse survival outcomes, comparable to human patients. The simplicity and scalability of the ptch1 MB model makes it highly amenable to CRISPR-based genome editing screens to identify genes required for SHH MB tumor formation in vivo, and here we identify the grk3 kinase as one such target.
Keywords: CRISPR; GRK2; GRK3; PTCH1; Pediatric brain tumors; SHH medulloblastoma; TP53; midbrain-hindbrain boundary; valvula cerebelli; zebrafish.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
