rCRUX: A Rapid and Versatile Tool for Generating Metabarcoding Reference libraries in R
- PMID: 38370872
- PMCID: PMC10871694
- DOI: 10.1002/edn3.489
rCRUX: A Rapid and Versatile Tool for Generating Metabarcoding Reference libraries in R
Abstract
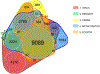
The sequencing revolution requires accurate taxonomic classification of DNA sequences. Key to making accurate taxonomic assignments are curated, comprehensive reference barcode databases. However, the generation and curation of such databases has remained challenging given the large and continuously growing volumes of both DNA sequence data and novel reference barcode targets. Monitoring and research applications require a greater diversity of specialized gene regions and targeted taxa then are currently curated by professional staff. Thus there is a growing need for an easy to implement computational tool that can generate comprehensive metabarcoding reference libraries for any bespoke locus. We address this need by reimagining CRUX from the Anacapa Toolkit and present the rCRUX package in R which, like it's predecessor, relies on sequence homology and PCR primer compatibility instead of keyword-searches to avoid limitations of user-defined metadata. The typical workflow involves searching for plausible seed amplicons (get_seeds_local() or get_seeds_remote()) by simulating in silico PCR to acquire a set of sequences analogous to PCR products containing a user-defined set of primer sequences. Next, these seeds are used to iteratively blast search seed sequences against a local copy of the National Center for Biotechnology Information (NCBI) formatted nt database using a taxonomic-rank based stratified random sampling approach ( blast_seeds() ). This results in a comprehensive set of sequence matches. This database is dereplicated and cleaned (derep_and_clean_db()) by identifying identical reference sequences and collapsing the taxonomic path to the lowest taxonomic agreement across all matching reads. This results in a curated, comprehensive database of primer-specific reference barcode sequences from NCBI. Databases can then be compared (compare_db()) to determine read and taxonomic overlap. We demonstrate that rCRUX provides more comprehensive reference databases for the MiFish Universal Teleost 12S, Taberlet trnl, fungal ITS, and Leray CO1 loci than CRABS, MetaCurator, RESCRIPt, and ecoPCR reference databases. We then further demonstrate the utility of rCRUX by generating 24 reference databases for 20 metabarcoding loci, many of which lack dedicated reference database curation efforts. The rCRUX package provides a simple to use tool for the generation of curated, comprehensive reference databases for user-defined loci, facilitating accurate and effective taxonomic classification of metabarcoding and DNA sequence efforts broadly.
Conflict of interest statement
Conflicts of Interest The authors have no conflict of interests to report.
Figures

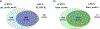



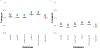
Update of
-
rCRUX: A Rapid and Versatile Tool for Generating Metabarcoding Reference libraries in R.bioRxiv [Preprint]. 2023 Jun 3:2023.05.31.543005. doi: 10.1101/2023.05.31.543005. bioRxiv. 2023. Update in: Environ DNA. 2024 Jan;6(1):e489. doi: 10.1002/edn3.489. PMID: 37397980 Free PMC article. Updated. Preprint.
Similar articles
-
rCRUX: A Rapid and Versatile Tool for Generating Metabarcoding Reference libraries in R.bioRxiv [Preprint]. 2023 Jun 3:2023.05.31.543005. doi: 10.1101/2023.05.31.543005. bioRxiv. 2023. Update in: Environ DNA. 2024 Jan;6(1):e489. doi: 10.1002/edn3.489. PMID: 37397980 Free PMC article. Updated. Preprint.
-
crabs-A software program to generate curated reference databases for metabarcoding sequencing data.Mol Ecol Resour. 2023 Apr;23(3):725-738. doi: 10.1111/1755-0998.13741. Epub 2022 Dec 11. Mol Ecol Resour. 2023. PMID: 36437603
-
taxalogue: a toolkit to create comprehensive CO1 reference databases.PeerJ. 2023 Dec 4;11:e16253. doi: 10.7717/peerj.16253. eCollection 2023. PeerJ. 2023. PMID: 38077427 Free PMC article.
-
Improving metabarcoding taxonomic assignment: A case study of fishes in a large marine ecosystem.Mol Ecol Resour. 2021 Oct;21(7):2546-2564. doi: 10.1111/1755-0998.13450. Epub 2021 Jul 8. Mol Ecol Resour. 2021. PMID: 34235858
-
DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work.Sci Total Environ. 2019 Aug 15;678:499-524. doi: 10.1016/j.scitotenv.2019.04.247. Epub 2019 Apr 27. Sci Total Environ. 2019. PMID: 31077928 Review.
Cited by
-
A new sampling device for metabarcoding surveillance of port communities and detection of non-indigenous species.iScience. 2023 Nov 25;27(1):108588. doi: 10.1016/j.isci.2023.108588. eCollection 2024 Jan 19. iScience. 2023. PMID: 38111684 Free PMC article.
-
Reference Sequence Browser: An R application with a user-friendly GUI to rapidly query sequence databases.PLoS One. 2024 Oct 31;19(10):e0309707. doi: 10.1371/journal.pone.0309707. eCollection 2024. PLoS One. 2024. PMID: 39480818 Free PMC article.
References
-
- Ahmed M, Back MA, Prior T, Karssen G, Lawson R, Adams I, & Sapp M. (2019). Metabarcoding of soil nematodes: the importance of taxonomic coverage and availability of reference sequences in choosing suitable marker (s). Metabarcoding and Metagenomics, 3, e36408.
-
- Asase A, Mzumara-Gawa TI, Owino JO, Peterson AT, & Saupe E. (2022). Replacing “parachute science” with “global science” in ecology and conservation biology. Conservation Science and Practice, 4(5), e517.
-
- Altschul SF, Gish W, Miller W, Myers EW, & Lipman DJ (1990). Basic local alignment search tool. Journal of molecular biology, 215(3), 403–410. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
