Phased genomics reveals hidden somatic mutations and provides insight into fruit development in sweet orange
- PMID: 38371640
- PMCID: PMC10873711
- DOI: 10.1093/hr/uhad268
Phased genomics reveals hidden somatic mutations and provides insight into fruit development in sweet orange
Abstract
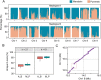
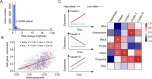
Although revisiting the discoveries and implications of genetic variations using phased genomics is critical, such efforts are still lacking. Somatic mutations represent a crucial source of genetic diversity for breeding and are especially remarkable in heterozygous perennial and asexual crops. In this study, we focused on a diploid sweet orange (Citrus sinensis) and constructed a haplotype-resolved genome using high fidelity (HiFi) reads, which revealed 10.6% new sequences. Based on the phased genome, we elucidate significant genetic admixtures and haplotype differences. We developed a somatic detection strategy that reveals hidden somatic mutations overlooked in a single reference genome. We generated a phased somatic variation map by combining high-depth whole-genome sequencing (WGS) data from 87 sweet orange somatic varieties. Notably, we found twice as many somatic mutations relative to a single reference genome. Using these hidden somatic mutations, we separated sweet oranges into seven major clades and provide insight into unprecedented genetic mosaicism and strong positive selection. Furthermore, these phased genomics data indicate that genomic heterozygous variations contribute to allele-specific expression during fruit development. By integrating allelic expression differences and somatic mutations, we identified a somatic mutation that induces increases in fruit size. Applications of phased genomics will lead to powerful approaches for discovering genetic variations and uncovering their effects in highly heterozygous plants. Our data provide insight into the hidden somatic mutation landscape in the sweet orange genome, which will facilitate citrus breeding.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
All authors have declared that no competing interests exist.
Figures


References
-
- D'Amato F. Role of somatic mutations in the evolution of higher plants. Caryologia. 1997;50:1–15
-
- Schoen DJ, Schultz ST. Somatic mutation and evolution in plants. Annu Rev Ecol Evol Syst. 2019;50:49–73
-
- Wang L, Huang Y, Liu ZA. et al. . Somatic variations led to the selection of acidic and acidless orange cultivars. Nat Plants. 2021;7:954–65 - PubMed
-
- Klekowski EJ Jr, Godfrey PJ. Ageing and mutation in plants. Nature. 1989;340:389–91
LinkOut - more resources
Full Text Sources

