Next-generation direct reprogramming
- PMID: 38371924
- PMCID: PMC10869521
- DOI: 10.3389/fcell.2024.1343106
Next-generation direct reprogramming
Abstract
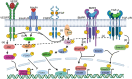
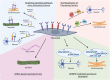
Tissue repair is significantly compromised in the aging human body resulting in critical disease conditions (such as myocardial infarction or Alzheimer's disease) and imposing a tremendous burden on global health. Reprogramming approaches (partial or direct reprogramming) are considered fruitful in addressing this unmet medical need. However, the efficacy, cellular maturity and specific targeting are still major challenges of direct reprogramming. Here we describe novel approaches in direct reprogramming that address these challenges. Extracellular signaling pathways (Receptor tyrosine kinases, RTK and Receptor Serine/Theronine Kinase, RSTK) and epigenetic marks remain central in rewiring the cellular program to determine the cell fate. We propose that modern protein design technologies (AI-designed minibinders regulating RTKs/RSTK, epigenetic enzymes, or pioneer factors) have potential to solve the aforementioned challenges. An efficient transdifferentiation/direct reprogramming may in the future provide molecular strategies to collectively reduce aging, fibrosis, and degenerative diseases.
Keywords: aging; cardiac muscles; direct reprogramming; partial reprogramming; pioneer factors; signaling; skeletal muscle; transdifferentiation.
Copyright © 2024 Keshri, Detraux, Phal, McCurdy, Jhajharia, Chan, Mathieu and Ruohola-Baker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

