Association between sleep-related phenotypes and gut microbiota: a two-sample bidirectional Mendelian randomization study
- PMID: 38371937
- PMCID: PMC10869596
- DOI: 10.3389/fmicb.2024.1341643
Association between sleep-related phenotypes and gut microbiota: a two-sample bidirectional Mendelian randomization study
Abstract
Background: An increasing body of evidence suggests a profound interrelation between the microbiome and sleep-related concerns. Nevertheless, current observational studies can merely establish their correlation, leaving causality unexplored.
Study objectives: To ascertain whether specific gut microbiota are causally linked to seven sleep-related characteristics and propose potential strategies for insomnia prevention.
Methods: The study employed an extensive dataset of gut microbiota genetic variations from the MiBioGen alliance, encompassing 18,340 individuals. Taxonomic classification was conducted, identifying 131 genera and 196 bacterial taxa for analysis. Sleep-related phenotype (SRP) data were sourced from the IEU OpenGWAS project, covering traits such as insomnia, chronotype, and snoring. Instrumental variables (IVs) were selected based on specific criteria, including locus-wide significance, linkage disequilibrium calculations, and allele frequency thresholds. Statistical methods were employed to explore causal relationships, including inverse variance weighted (IVW), MR-Egger, weighted median, and weighted Mode. Sensitivity analyses, pleiotropy assessments, and Bonferroni corrections ensured result validity. Reverse causality analysis and adherence to STROBE-MR guidelines were conducted to bolster the study's rigor.
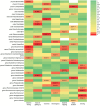
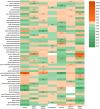
Results: Bidirectional Mendelian randomization (MR) analysis reveals a causative interplay between selected gut microbiota and sleep-related phenotypes. Notably, outcomes from the rigorously Bonferroni-corrected examination illuminate profound correlations amid precise compositions of the intestinal microbiome and slumber-associated parameters. Elevated abundance within the taxonomic ranks of class Negativicutes and order Selenomonadales was markedly associated with heightened susceptibility to severe insomnia (OR = 1.03, 95% CI: 1.02-1.05, p = 0.0001). Conversely, the augmented representation of the phylum Lentisphaerae stands in concord with protracted sleep duration (OR = 1.02, 95% CI: 1.01-1.04, p = 0.0005). Furthermore, heightened exposure to the genus Senegalimassilia exhibits the potential to ameliorate the manifestation of snoring symptoms (OR = 0.98, 95% CI: 0.96-0.99, p = 0.0001).
Conclusion: This study has unveiled the causal relationship between gut microbiota and SRPs, bestowing significant latent value upon future endeavors in both foundational research and clinical therapy.
Keywords: Mendelian randomization; causal effect; gut microbiota; insomnia; sleep-related phenotypes.
Copyright © 2024 Wang, Wang, Liu, Wan, Wu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Causal Effects of Gut Microbiota on Sleep-Related Phenotypes: A Two-Sample Mendelian Randomization Study.Clocks Sleep. 2023 Sep 12;5(3):566-580. doi: 10.3390/clockssleep5030037. Clocks Sleep. 2023. PMID: 37754355 Free PMC article.
-
Causal relationship between the gut microbiota and insomnia: a two-sample Mendelian randomization study.Front Cell Infect Microbiol. 2024 Mar 4;14:1279218. doi: 10.3389/fcimb.2024.1279218. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38500501 Free PMC article.
-
Causal relationship between gut microbiota and tuberculosis: a bidirectional two-sample Mendelian randomization analysis.Respir Res. 2024 Jan 4;25(1):16. doi: 10.1186/s12931-023-02652-7. Respir Res. 2024. PMID: 38178098 Free PMC article.
-
Exploring the potential causal relationship between gut microbiota and heart failure: A two-sample mendelian randomization study combined with the geo database.Curr Probl Cardiol. 2024 Feb;49(2):102235. doi: 10.1016/j.cpcardiol.2023.102235. Epub 2023 Nov 30. Curr Probl Cardiol. 2024. PMID: 38040216 Review.
-
Causal relationship between gut microbiota and polycystic ovary syndrome: a literature review and Mendelian randomization study.Front Endocrinol (Lausanne). 2024 Feb 1;15:1280983. doi: 10.3389/fendo.2024.1280983. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38362275 Free PMC article. Review.
Cited by
-
Causal Associations Between Sleep Traits and Delirium: A Bidirectional Two-Sample Mendelian Randomization Study.Nat Sci Sleep. 2024 Dec 21;16:2171-2181. doi: 10.2147/NSS.S491216. eCollection 2024. Nat Sci Sleep. 2024. PMID: 39726858 Free PMC article.
-
Better objective sleep quality is associated with higher gut microbiota richness in older adults.Geroscience. 2025 Jun;47(3):4121-4137. doi: 10.1007/s11357-025-01524-w. Epub 2025 Jan 31. Geroscience. 2025. PMID: 39888583 Free PMC article.
-
Banxia-Yiyiren alleviates insomnia and anxiety by regulating the gut microbiota and metabolites of PCPA-induced insomnia model rats.Front Microbiol. 2024 Nov 7;15:1405566. doi: 10.3389/fmicb.2024.1405566. eCollection 2024. Front Microbiol. 2024. PMID: 39575182 Free PMC article.
-
Lactobacillaceae-mediated eye-brain-gut axis regulates high myopia-related anxiety: from the perspective of predictive, preventive, and personalized medicine.EPMA J. 2024 Nov 26;15(4):573-585. doi: 10.1007/s13167-024-00387-z. eCollection 2024 Dec. EPMA J. 2024. PMID: 39635020
References
-
- Benedict C., Vogel H., Jonas W., Woting A., Blaut M., Schürmann A., et al. . (2016). Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Mol Metab. 5, 1175–1186. doi: 10.1016/j.molmet.2016.10.003, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

