Spatially resolved transcriptome of the aging mouse brain
- PMID: 38372175
- PMCID: PMC11113349
- DOI: 10.1111/acel.14109
Spatially resolved transcriptome of the aging mouse brain
Abstract
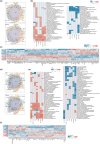
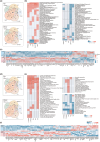
Brain aging is associated with cognitive decline, memory loss and many neurodegenerative disorders. The mammalian brain has distinct structural regions that perform specific functions. However, our understanding in gene expression and cell types within the context of the spatial organization of the mammalian aging brain is limited. Here we generated spatial transcriptomic maps of young and old mouse brains. We identified 27 distinguished brain spatial domains, including layer-specific subregions that are difficult to dissect individually. We comprehensively characterized spatial-specific changes in gene expression in the aging brain, particularly for isocortex, the hippocampal formation, brainstem and fiber tracts, and validated some gene expression differences by qPCR and immunohistochemistry. We identified aging-related genes and pathways that vary in a coordinated manner across spatial regions and parsed the spatial features of aging-related signals, providing important clues to understand genes with specific functions in different brain regions during aging. Combined with single-cell transcriptomics data, we characterized the spatial distribution of brain cell types. The proportion of immature neurons decreased in the DG region with aging, indicating that the formation of new neurons is blocked. Finally, we detected changes in information interactions between regions and found specific pathways were deregulated with aging, including classic signaling WNT and layer-specific signaling COLLAGEN. In summary, we established a spatial molecular atlas of the aging mouse brain (http://sysbio.gzzoc.com/Mouse-Brain-Aging/), which provides important resources and novel insights into the molecular mechanism of brain aging.
Keywords: aging; brain; cell types; gene expression; spatial transcriptome.
© 2024 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare no competing financial interests.
Figures





References
-
- Berchtold, N. C. , Coleman, P. D. , Cribbs, D. H. , Rogers, J. , Gillen, D. L. , & Cotman, C. W. (2013). Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer's disease. Neurobiology of Aging, 34(6), 1653–1661. 10.1016/j.neurobiolaging.2012.11.024 - DOI - PMC - PubMed
-
- Berchtold, N. C. , Cribbs, D. H. , Coleman, P. D. , Rogers, J. , Head, E. , Kim, R. , Beach, T. , Miller, C. , Troncoso, J. , Trojanowski, J. Q. , Zielke, H. R. , & Cotman, C. W. (2008). Gene expression changes in the course of normal brain aging are sexually dimorphic. Proceedings of the National Academy of Sciences of the United States of America, 105(40), 15605–15610. 10.1073/pnas.0806883105 - DOI - PMC - PubMed
-
- Bouhrara, M. , Cortina, L. E. , Rejimon, A. C. , Khattar, N. , Bergeron, C. , Bergeron, J. , Melvin, D. , Zukley, L. , & Spencer, R. G. (2020). Quantitative age‐dependent differences in human brainstem myelination assessed using high‐resolution magnetic resonance mapping. NeuroImage, 206, 116307. 10.1016/j.neuroimage.2019.116307 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 2021YFF1200903/National Key R&D Program of China
- 202201020336/Guangzhou Basic and Applied Basic Research Foundation
- 82171404/National Natural Science Foundation of China
- 2022YFF1202901/The Key Research and Development Program of the Ministry of Science and Technology
- 2023A1515011529/Natural Science Foundation of Guangdong Province
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

