The genome and transcriptome of the snail Biomphalaria sudanica s.l.: immune gene diversification and highly polymorphic genomic regions in an important African vector of Schistosoma mansoni
- PMID: 38373909
- PMCID: PMC10875847
- DOI: 10.1186/s12864-024-10103-w
The genome and transcriptome of the snail Biomphalaria sudanica s.l.: immune gene diversification and highly polymorphic genomic regions in an important African vector of Schistosoma mansoni
Abstract
Background: Control and elimination of schistosomiasis is an arduous task, with current strategies proving inadequate to break transmission. Exploration of genetic approaches to interrupt Schistosoma mansoni transmission, the causative agent for human intestinal schistosomiasis in sub-Saharan Africa and South America, has led to genomic research of the snail vector hosts of the genus Biomphalaria. Few complete genomic resources exist, with African Biomphalaria species being particularly underrepresented despite this being where the majority of S. mansoni infections occur. Here we generate and annotate the first genome assembly of Biomphalaria sudanica sensu lato, a species responsible for S. mansoni transmission in lake and marsh habitats of the African Rift Valley. Supported by whole-genome diversity data among five inbred lines, we describe orthologs of immune-relevant gene regions in the South American vector B. glabrata and present a bioinformatic pipeline to identify candidate novel pathogen recognition receptors (PRRs).
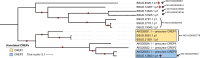
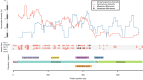
Results: De novo genome and transcriptome assembly of inbred B. sudanica originating from the shoreline of Lake Victoria (Kisumu, Kenya) resulted in a haploid genome size of ~ 944.2 Mb (6,728 fragments, N50 = 1.067 Mb), comprising 23,598 genes (BUSCO = 93.6% complete). The B. sudanica genome contains orthologues to all described immune genes/regions tied to protection against S. mansoni in B. glabrata, including the polymorphic transmembrane clusters (PTC1 and PTC2), RADres, and other loci. The B. sudanica PTC2 candidate immune genomic region contained many PRR-like genes across a much wider genomic region than has been shown in B. glabrata, as well as a large inversion between species. High levels of intra-species nucleotide diversity were seen in PTC2, as well as in regions linked to PTC1 and RADres orthologues. Immune related and putative PRR gene families were significantly over-represented in the sub-set of B. sudanica genes determined as hyperdiverse, including high extracellular diversity in transmembrane genes, which could be under pathogen-mediated balancing selection. However, no overall expansion in immunity related genes was seen in African compared to South American lineages.
Conclusions: The B. sudanica genome and analyses presented here will facilitate future research in vector immune defense mechanisms against pathogens. This genomic/transcriptomic resource provides necessary data for the future development of molecular snail vector control/surveillance tools, facilitating schistosome transmission interruption mechanisms in Africa.
Keywords: Balancing selection; Biomphalaria choanomphala; Biomphalaria sudanica; De novo genome assembly; Gene family evolution; Immunogenetics; Pathogen recognition; Polymorphism; Schistosomiasis; Snail vector.
© 2024. The Author(s).
Conflict of interest statement
Not applicable.
Figures








Update of
-
The genome and transcriptome of the snail Biomphalaria sudanica s.l.: Immune gene diversification and highly polymorphic genomic regions in an important African vector of Schistosoma mansoni.bioRxiv [Preprint]. 2023 Nov 2:2023.11.01.565203. doi: 10.1101/2023.11.01.565203. bioRxiv. 2023. Update in: BMC Genomics. 2024 Feb 19;25(1):192. doi: 10.1186/s12864-024-10103-w. PMID: 37961413 Free PMC article. Updated. Preprint.
References
-
- WHO. Schistosomiasis: Key facts. 2023 [cited 2023 Apr 14]. Available from: https://www.who.int/news-room/fact-sheets/detail/schistosomiasis.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

