A transient protein folding response targets aggregation in the early phase of TDP-43-mediated neurodegeneration
- PMID: 38374041
- PMCID: PMC10876645
- DOI: 10.1038/s41467-024-45646-9
A transient protein folding response targets aggregation in the early phase of TDP-43-mediated neurodegeneration
Abstract
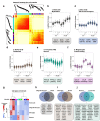
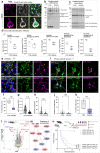
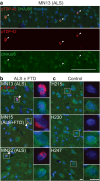
Understanding the mechanisms that drive TDP-43 pathology is integral to combating amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD) and other neurodegenerative diseases. Here we generated a longitudinal quantitative proteomic map of the cortex from the cytoplasmic TDP-43 rNLS8 mouse model of ALS and FTLD, and developed a complementary open-access webtool, TDP-map ( https://shiny.rcc.uq.edu.au/TDP-map/ ). We identified distinct protein subsets enriched for diverse biological pathways with temporal alterations in protein abundance, including increases in protein folding factors prior to disease onset. This included increased levels of DnaJ homolog subfamily B member 5, DNAJB5, which also co-localized with TDP-43 pathology in diseased human motor cortex. DNAJB5 over-expression decreased TDP-43 aggregation in cell and cortical neuron cultures, and knockout of Dnajb5 exacerbated motor impairments caused by AAV-mediated cytoplasmic TDP-43 expression in mice. Together, these findings reveal molecular mechanisms at distinct stages of ALS and FTLD progression and suggest that protein folding factors could be protective in neurodegenerative diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

