PIBF1 regulates trophoblast syncytialization and promotes cardiovascular development
- PMID: 38374152
- PMCID: PMC10876648
- DOI: 10.1038/s41467-024-45647-8
PIBF1 regulates trophoblast syncytialization and promotes cardiovascular development
Abstract
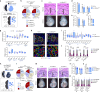
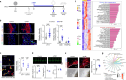
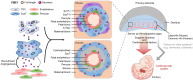
Proper placental development in early pregnancy ensures a positive outcome later on. The developmental relationship between the placenta and embryonic organs, such as the heart, is crucial for a normal pregnancy. However, the mechanism through which the placenta influences the development of embryonic organs remains unclear. Trophoblasts fuse to form multinucleated syncytiotrophoblasts (SynT), which primarily make up the placental materno-fetal interface. We discovered that endogenous progesterone immunomodulatory binding factor 1 (PIBF1) is vital for trophoblast differentiation and fusion into SynT in humans and mice. PIBF1 facilitates communication between SynT and adjacent vascular cells, promoting vascular network development in the primary placenta. This process affected the early development of the embryonic cardiovascular system in mice. Moreover, in vitro experiments showed that PIBF1 promotes the development of cardiovascular characteristics in heart organoids. Our findings show how SynTs organize the barrier and imply their possible roles in supporting embryogenesis, including cardiovascular development. SynT-derived factors and SynT within the placenta may play critical roles in ensuring proper organogenesis of other organs in the embryo.
© 2024. The Author(s).
Conflict of interest statement
J.G.L., J.-M.Y., K.K., Y.S.C., and I.-J.B. have filed a provisional patent application in the Korea patent office (composition for preventing and treating pregnant complications related to placenta and/or fetal cardiovascular abnormalities by targeting PIBF1). All the other authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- 2020R1A2C2012912/National Research Foundation of Korea (NRF)
- 2018M3A9D5A01087977/National Research Foundation of Korea (NRF)
- 2020-028/Asan Institute for Life Sciences, Asan Medical Center
- 2022-013/Asan Institute for Life Sciences, Asan Medical Center
- 2023-062/Asan Institute for Life Sciences, Asan Medical Center
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

